
Marine snail associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
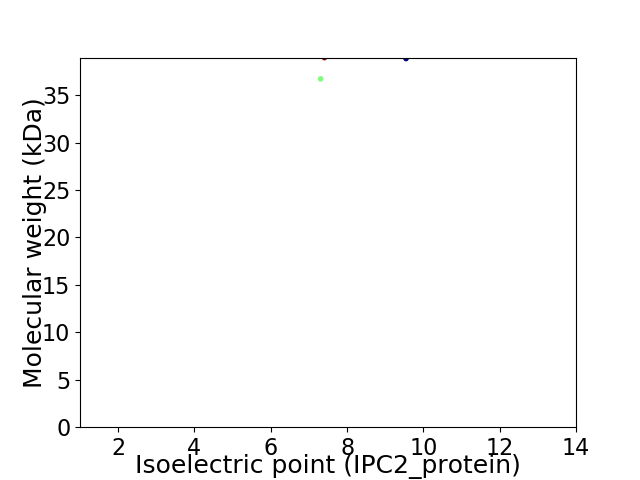
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL52|A0A0K1RL52_9CIRC Putative replication initiation protein OS=Marine snail associated circular virus OX=1692255 PE=4 SV=1
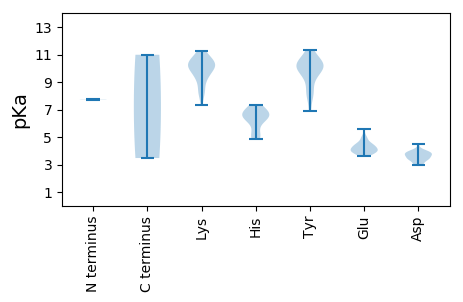
MM1 pKa = 7.71AHH3 pKa = 6.5RR4 pKa = 11.84QHH6 pKa = 6.86AAKK9 pKa = 10.11RR10 pKa = 11.84WVFTINNWTAAEE22 pKa = 3.96QQALIDD28 pKa = 3.79SSDD31 pKa = 3.44NFDD34 pKa = 3.69YY35 pKa = 11.36LCFGRR40 pKa = 11.84EE41 pKa = 3.63RR42 pKa = 11.84GDD44 pKa = 3.23NNTPHH49 pKa = 6.27LQGYY53 pKa = 9.13VILKK57 pKa = 8.72TKK59 pKa = 10.62LRR61 pKa = 11.84LNNVKK66 pKa = 10.15ALPGFRR72 pKa = 11.84RR73 pKa = 11.84CHH75 pKa = 6.9LEE77 pKa = 3.72VSRR80 pKa = 11.84GTPQQAADD88 pKa = 3.74YY89 pKa = 9.05CKK91 pKa = 10.44KK92 pKa = 10.81DD93 pKa = 2.99GDD95 pKa = 3.88FEE97 pKa = 4.73EE98 pKa = 5.61FGEE101 pKa = 4.35LPKK104 pKa = 11.19GQGKK108 pKa = 10.23RR109 pKa = 11.84SDD111 pKa = 3.71FEE113 pKa = 4.4NLKK116 pKa = 10.35EE117 pKa = 4.2WIKK120 pKa = 11.26SLDD123 pKa = 3.38HH124 pKa = 6.72WPDD127 pKa = 2.98DD128 pKa = 3.66HH129 pKa = 7.23EE130 pKa = 4.42IAEE133 pKa = 4.91EE134 pKa = 4.46YY135 pKa = 9.61PSLWGRR141 pKa = 11.84YY142 pKa = 7.85RR143 pKa = 11.84SACEE147 pKa = 3.67SFRR150 pKa = 11.84QLFGKK155 pKa = 9.32PIEE158 pKa = 4.5VVDD161 pKa = 3.78PTTFEE166 pKa = 3.86PRR168 pKa = 11.84VWQQRR173 pKa = 11.84IIDD176 pKa = 3.96ICNGEE181 pKa = 3.93PDD183 pKa = 3.42PRR185 pKa = 11.84KK186 pKa = 10.01VYY188 pKa = 10.24FVVDD192 pKa = 3.75EE193 pKa = 4.58NGNTGKK199 pKa = 10.54SYY201 pKa = 11.16LSAYY205 pKa = 9.81LISKK209 pKa = 10.1FPDD212 pKa = 2.96EE213 pKa = 4.22VQILSVGRR221 pKa = 11.84RR222 pKa = 11.84DD223 pKa = 4.49DD224 pKa = 3.66LAHH227 pKa = 7.35AINPRR232 pKa = 11.84RR233 pKa = 11.84SIFLFDD239 pKa = 3.6VPRR242 pKa = 11.84GGMEE246 pKa = 3.76YY247 pKa = 10.24LQYY250 pKa = 10.96TIFEE254 pKa = 4.16QLKK257 pKa = 8.49NRR259 pKa = 11.84TVFSPKK265 pKa = 9.81YY266 pKa = 10.25NSITKK271 pKa = 10.05ILRR274 pKa = 11.84KK275 pKa = 9.29LPHH278 pKa = 6.31VIIFSNEE285 pKa = 3.77SPDD288 pKa = 3.81RR289 pKa = 11.84NKK291 pKa = 10.0MSHH294 pKa = 6.57DD295 pKa = 3.97RR296 pKa = 11.84YY297 pKa = 9.97HH298 pKa = 5.43VTHH301 pKa = 6.79IRR303 pKa = 11.84QFPRR307 pKa = 11.84ADD309 pKa = 3.4DD310 pKa = 3.81NGANGG315 pKa = 3.47
MM1 pKa = 7.71AHH3 pKa = 6.5RR4 pKa = 11.84QHH6 pKa = 6.86AAKK9 pKa = 10.11RR10 pKa = 11.84WVFTINNWTAAEE22 pKa = 3.96QQALIDD28 pKa = 3.79SSDD31 pKa = 3.44NFDD34 pKa = 3.69YY35 pKa = 11.36LCFGRR40 pKa = 11.84EE41 pKa = 3.63RR42 pKa = 11.84GDD44 pKa = 3.23NNTPHH49 pKa = 6.27LQGYY53 pKa = 9.13VILKK57 pKa = 8.72TKK59 pKa = 10.62LRR61 pKa = 11.84LNNVKK66 pKa = 10.15ALPGFRR72 pKa = 11.84RR73 pKa = 11.84CHH75 pKa = 6.9LEE77 pKa = 3.72VSRR80 pKa = 11.84GTPQQAADD88 pKa = 3.74YY89 pKa = 9.05CKK91 pKa = 10.44KK92 pKa = 10.81DD93 pKa = 2.99GDD95 pKa = 3.88FEE97 pKa = 4.73EE98 pKa = 5.61FGEE101 pKa = 4.35LPKK104 pKa = 11.19GQGKK108 pKa = 10.23RR109 pKa = 11.84SDD111 pKa = 3.71FEE113 pKa = 4.4NLKK116 pKa = 10.35EE117 pKa = 4.2WIKK120 pKa = 11.26SLDD123 pKa = 3.38HH124 pKa = 6.72WPDD127 pKa = 2.98DD128 pKa = 3.66HH129 pKa = 7.23EE130 pKa = 4.42IAEE133 pKa = 4.91EE134 pKa = 4.46YY135 pKa = 9.61PSLWGRR141 pKa = 11.84YY142 pKa = 7.85RR143 pKa = 11.84SACEE147 pKa = 3.67SFRR150 pKa = 11.84QLFGKK155 pKa = 9.32PIEE158 pKa = 4.5VVDD161 pKa = 3.78PTTFEE166 pKa = 3.86PRR168 pKa = 11.84VWQQRR173 pKa = 11.84IIDD176 pKa = 3.96ICNGEE181 pKa = 3.93PDD183 pKa = 3.42PRR185 pKa = 11.84KK186 pKa = 10.01VYY188 pKa = 10.24FVVDD192 pKa = 3.75EE193 pKa = 4.58NGNTGKK199 pKa = 10.54SYY201 pKa = 11.16LSAYY205 pKa = 9.81LISKK209 pKa = 10.1FPDD212 pKa = 2.96EE213 pKa = 4.22VQILSVGRR221 pKa = 11.84RR222 pKa = 11.84DD223 pKa = 4.49DD224 pKa = 3.66LAHH227 pKa = 7.35AINPRR232 pKa = 11.84RR233 pKa = 11.84SIFLFDD239 pKa = 3.6VPRR242 pKa = 11.84GGMEE246 pKa = 3.76YY247 pKa = 10.24LQYY250 pKa = 10.96TIFEE254 pKa = 4.16QLKK257 pKa = 8.49NRR259 pKa = 11.84TVFSPKK265 pKa = 9.81YY266 pKa = 10.25NSITKK271 pKa = 10.05ILRR274 pKa = 11.84KK275 pKa = 9.29LPHH278 pKa = 6.31VIIFSNEE285 pKa = 3.77SPDD288 pKa = 3.81RR289 pKa = 11.84NKK291 pKa = 10.0MSHH294 pKa = 6.57DD295 pKa = 3.97RR296 pKa = 11.84YY297 pKa = 9.97HH298 pKa = 5.43VTHH301 pKa = 6.79IRR303 pKa = 11.84QFPRR307 pKa = 11.84ADD309 pKa = 3.4DD310 pKa = 3.81NGANGG315 pKa = 3.47
Molecular weight: 36.71 kDa
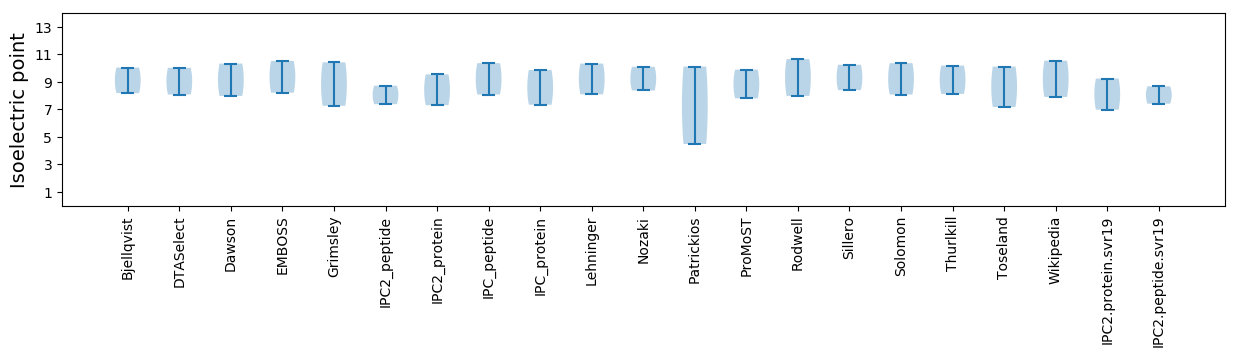
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL52|A0A0K1RL52_9CIRC Putative replication initiation protein OS=Marine snail associated circular virus OX=1692255 PE=4 SV=1
MM1 pKa = 7.73SFEE4 pKa = 4.25TAVTAGFVAGYY15 pKa = 9.98GYY17 pKa = 10.0VARR20 pKa = 11.84EE21 pKa = 4.11TYY23 pKa = 10.12RR24 pKa = 11.84ALQEE28 pKa = 4.2GRR30 pKa = 11.84SQSMYY35 pKa = 10.34HH36 pKa = 5.17YY37 pKa = 10.27NLRR40 pKa = 11.84KK41 pKa = 9.6RR42 pKa = 11.84KK43 pKa = 8.06PSAVITPSKK52 pKa = 10.62APVIRR57 pKa = 11.84RR58 pKa = 11.84TAPTQAPEE66 pKa = 3.94MRR68 pKa = 11.84GLTVHH73 pKa = 6.39TRR75 pKa = 11.84TTRR78 pKa = 11.84PWRR81 pKa = 11.84TSNYY85 pKa = 6.86RR86 pKa = 11.84TLGRR90 pKa = 11.84RR91 pKa = 11.84PGKK94 pKa = 9.44YY95 pKa = 9.58GSRR98 pKa = 11.84KK99 pKa = 8.17RR100 pKa = 11.84TISADD105 pKa = 3.42NTSSAEE111 pKa = 3.98KK112 pKa = 9.73MPSDD116 pKa = 3.94KK117 pKa = 10.91SVSADD122 pKa = 2.95GLIFVEE128 pKa = 4.55QQSTTSQINKK138 pKa = 9.24RR139 pKa = 11.84RR140 pKa = 11.84GEE142 pKa = 4.0LVNIRR147 pKa = 11.84GVKK150 pKa = 9.22VRR152 pKa = 11.84YY153 pKa = 8.17FFKK156 pKa = 10.37MNQRR160 pKa = 11.84LNPTNAITIPVRR172 pKa = 11.84VRR174 pKa = 11.84WAIIIPKK181 pKa = 10.01ADD183 pKa = 3.67TLTLTTVGTAPNSYY197 pKa = 10.39SRR199 pKa = 11.84PALPNFFINEE209 pKa = 3.99NPVDD213 pKa = 4.07RR214 pKa = 11.84VAKK217 pKa = 10.67DD218 pKa = 3.31FDD220 pKa = 3.99TSVPNPGNHH229 pKa = 4.88WDD231 pKa = 3.48MFNRR235 pKa = 11.84KK236 pKa = 8.31INRR239 pKa = 11.84EE240 pKa = 3.94EE241 pKa = 3.97YY242 pKa = 10.28SVLKK246 pKa = 10.15EE247 pKa = 4.02GQFILCQDD255 pKa = 3.93GGNGEE260 pKa = 4.3LSGIRR265 pKa = 11.84PRR267 pKa = 11.84PMRR270 pKa = 11.84TYY272 pKa = 10.6KK273 pKa = 10.47HH274 pKa = 6.31LNFYY278 pKa = 10.59IPIKK282 pKa = 9.98RR283 pKa = 11.84QMHH286 pKa = 4.84FTGTGGEE293 pKa = 4.1PDD295 pKa = 3.49ANVYY299 pKa = 8.34MLYY302 pKa = 9.68WYY304 pKa = 10.43AFEE307 pKa = 4.99GSNAATKK314 pKa = 10.41DD315 pKa = 3.1IADD318 pKa = 3.97NADD321 pKa = 3.31KK322 pKa = 10.91PFVEE326 pKa = 5.02FHH328 pKa = 6.7EE329 pKa = 4.43KK330 pKa = 7.34TTYY333 pKa = 8.21FTNSDD338 pKa = 3.42MFKK341 pKa = 10.98
MM1 pKa = 7.73SFEE4 pKa = 4.25TAVTAGFVAGYY15 pKa = 9.98GYY17 pKa = 10.0VARR20 pKa = 11.84EE21 pKa = 4.11TYY23 pKa = 10.12RR24 pKa = 11.84ALQEE28 pKa = 4.2GRR30 pKa = 11.84SQSMYY35 pKa = 10.34HH36 pKa = 5.17YY37 pKa = 10.27NLRR40 pKa = 11.84KK41 pKa = 9.6RR42 pKa = 11.84KK43 pKa = 8.06PSAVITPSKK52 pKa = 10.62APVIRR57 pKa = 11.84RR58 pKa = 11.84TAPTQAPEE66 pKa = 3.94MRR68 pKa = 11.84GLTVHH73 pKa = 6.39TRR75 pKa = 11.84TTRR78 pKa = 11.84PWRR81 pKa = 11.84TSNYY85 pKa = 6.86RR86 pKa = 11.84TLGRR90 pKa = 11.84RR91 pKa = 11.84PGKK94 pKa = 9.44YY95 pKa = 9.58GSRR98 pKa = 11.84KK99 pKa = 8.17RR100 pKa = 11.84TISADD105 pKa = 3.42NTSSAEE111 pKa = 3.98KK112 pKa = 9.73MPSDD116 pKa = 3.94KK117 pKa = 10.91SVSADD122 pKa = 2.95GLIFVEE128 pKa = 4.55QQSTTSQINKK138 pKa = 9.24RR139 pKa = 11.84RR140 pKa = 11.84GEE142 pKa = 4.0LVNIRR147 pKa = 11.84GVKK150 pKa = 9.22VRR152 pKa = 11.84YY153 pKa = 8.17FFKK156 pKa = 10.37MNQRR160 pKa = 11.84LNPTNAITIPVRR172 pKa = 11.84VRR174 pKa = 11.84WAIIIPKK181 pKa = 10.01ADD183 pKa = 3.67TLTLTTVGTAPNSYY197 pKa = 10.39SRR199 pKa = 11.84PALPNFFINEE209 pKa = 3.99NPVDD213 pKa = 4.07RR214 pKa = 11.84VAKK217 pKa = 10.67DD218 pKa = 3.31FDD220 pKa = 3.99TSVPNPGNHH229 pKa = 4.88WDD231 pKa = 3.48MFNRR235 pKa = 11.84KK236 pKa = 8.31INRR239 pKa = 11.84EE240 pKa = 3.94EE241 pKa = 3.97YY242 pKa = 10.28SVLKK246 pKa = 10.15EE247 pKa = 4.02GQFILCQDD255 pKa = 3.93GGNGEE260 pKa = 4.3LSGIRR265 pKa = 11.84PRR267 pKa = 11.84PMRR270 pKa = 11.84TYY272 pKa = 10.6KK273 pKa = 10.47HH274 pKa = 6.31LNFYY278 pKa = 10.59IPIKK282 pKa = 9.98RR283 pKa = 11.84QMHH286 pKa = 4.84FTGTGGEE293 pKa = 4.1PDD295 pKa = 3.49ANVYY299 pKa = 8.34MLYY302 pKa = 9.68WYY304 pKa = 10.43AFEE307 pKa = 4.99GSNAATKK314 pKa = 10.41DD315 pKa = 3.1IADD318 pKa = 3.97NADD321 pKa = 3.31KK322 pKa = 10.91PFVEE326 pKa = 5.02FHH328 pKa = 6.7EE329 pKa = 4.43KK330 pKa = 7.34TTYY333 pKa = 8.21FTNSDD338 pKa = 3.42MFKK341 pKa = 10.98
Molecular weight: 38.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656 |
315 |
341 |
328.0 |
37.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.098 ± 0.739 | 0.915 ± 0.488 |
5.64 ± 1.206 | 5.488 ± 0.625 |
5.335 ± 0.275 | 6.25 ± 0.158 |
2.591 ± 0.654 | 5.64 ± 0.284 |
5.945 ± 0.063 | 5.64 ± 0.976 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.982 ± 0.747 | 6.098 ± 0.278 |
6.098 ± 0.278 | 3.659 ± 0.571 |
8.689 ± 0.316 | 6.098 ± 0.278 |
6.555 ± 1.993 | 5.488 ± 0.297 |
1.524 ± 0.276 | 4.268 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
