
Leptothrix ochracea L12
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiales genera incertae sedis; Leptothrix; Leptothrix ochracea
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
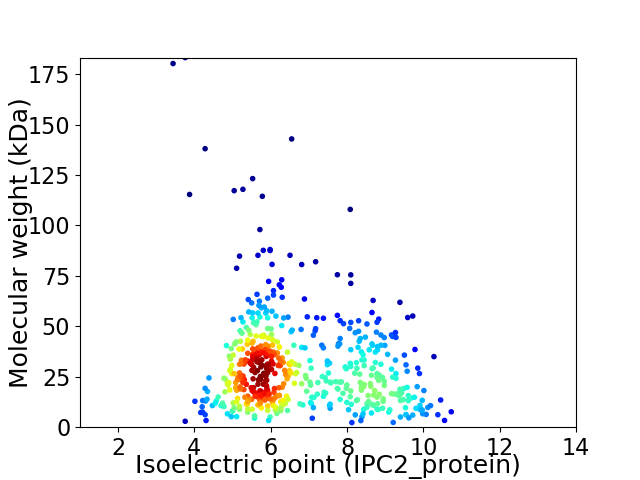
Virtual 2D-PAGE plot for 531 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
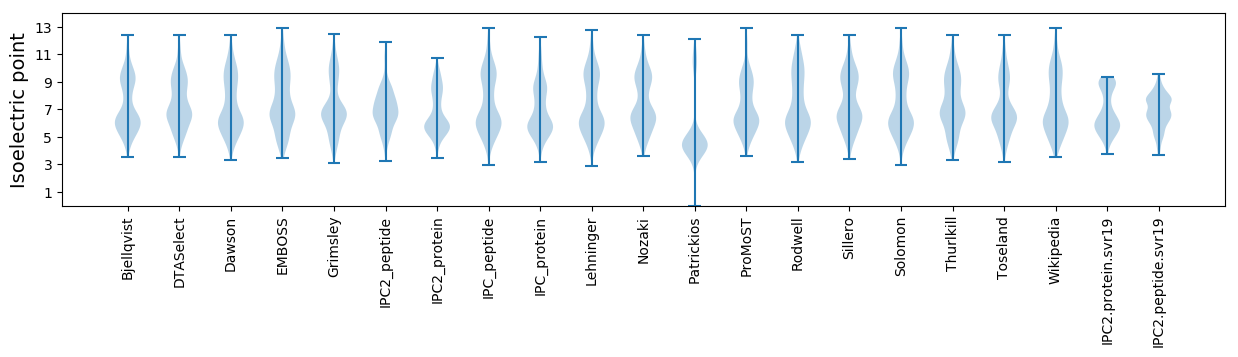
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4Z631|I4Z631_9BURK GDT1 family protein OS=Leptothrix ochracea L12 OX=735332 GN=LepocDRAFT_00004070 PE=3 SV=1
MM1 pKa = 7.44NEE3 pKa = 3.95STVNPSVMPAIGPTQQGAGVNHH25 pKa = 7.43GGLSITPQRR34 pKa = 11.84HH35 pKa = 5.26EE36 pKa = 4.24GHH38 pKa = 6.37PTAAVTPAPLAHH50 pKa = 7.34RR51 pKa = 11.84FGQAPDD57 pKa = 3.68TEE59 pKa = 4.6GTEE62 pKa = 4.22EE63 pKa = 5.61DD64 pKa = 4.4SIATNAVEE72 pKa = 4.24TVEE75 pKa = 4.43TLDD78 pKa = 3.87EE79 pKa = 4.3DD80 pKa = 4.25TFGADD85 pKa = 3.06ISDD88 pKa = 3.63GTEE91 pKa = 3.75AFDD94 pKa = 4.5PGTMGHH100 pKa = 6.19EE101 pKa = 4.09SSGRR105 pKa = 11.84EE106 pKa = 3.56DD107 pKa = 3.48TLRR110 pKa = 11.84YY111 pKa = 9.37SALHH115 pKa = 6.35EE116 pKa = 4.2PAMMLTAADD125 pKa = 3.67ATFIASDD132 pKa = 3.95PAPLALAQPMEE143 pKa = 4.32YY144 pKa = 10.06LGSSAAASGGSDD156 pKa = 2.75AGAAAVGSTAMGEE169 pKa = 4.11LMPMLGAVTFGVIAATSANNATQAQTPNPSPTATGTAPPLVFSISHH215 pKa = 6.48AAGTPHH221 pKa = 7.49VMVVDD226 pKa = 3.8VTLPSTMVAGQTLTFSVAQGNAVLEE251 pKa = 4.4TVNYY255 pKa = 10.44VLTQTDD261 pKa = 3.47INAGSITKK269 pKa = 10.02FLGSNNLSDD278 pKa = 3.9SNYY281 pKa = 10.1QISVNQAGTTALASNTSTFHH301 pKa = 7.52LDD303 pKa = 2.98VTAPSAPQLNLSPNGNGTTVNVSLPIDD330 pKa = 3.72AVVGDD335 pKa = 4.68SLTLTLRR342 pKa = 11.84DD343 pKa = 3.57QTGSAVQTVTQAVTALNLSTHH364 pKa = 6.04TLQLSLNTGALPANTSYY381 pKa = 11.02QALANLTDD389 pKa = 3.22VSGNVSINSTPISMTTPGAPTPGITLTQDD418 pKa = 2.43SGTPGDD424 pKa = 4.73RR425 pKa = 11.84ITNQAGISLSGVLSSGVVEE444 pKa = 4.03YY445 pKa = 10.94SVDD448 pKa = 3.26GGAWTSVYY456 pKa = 8.83TAPTQNGNHH465 pKa = 5.3TVQVRR470 pKa = 11.84QFDD473 pKa = 4.28TNTGHH478 pKa = 6.32VSNPGSLSFTLDD490 pKa = 3.06TLAPAALIPVLANGANGLTNSAALAALTPEE520 pKa = 4.06VGALLEE526 pKa = 3.96YY527 pKa = 10.45RR528 pKa = 11.84VDD530 pKa = 3.46GGAWASSYY538 pKa = 7.73TAPTGDD544 pKa = 3.12IAHH547 pKa = 6.17TVDD550 pKa = 4.48VRR552 pKa = 11.84QTDD555 pKa = 3.27AAGNISAWFTLSFTLDD571 pKa = 3.24TVAPASVTPTLANGATTLTNNAALTALSATEE602 pKa = 3.78VGATVEE608 pKa = 3.97YY609 pKa = 10.42RR610 pKa = 11.84VDD612 pKa = 3.24GGAWGAYY619 pKa = 5.79TAPSAQGAHH628 pKa = 4.7TVEE631 pKa = 4.02ARR633 pKa = 11.84QVDD636 pKa = 3.65AAGNASAASAPLAFTLDD653 pKa = 3.39TQAPVAPGVALSNGNPAGSTTNNGSVSVTGIEE685 pKa = 3.84AGATVEE691 pKa = 4.06YY692 pKa = 10.23QINASGTWLPLVADD706 pKa = 3.84AQGAYY711 pKa = 9.52NVPSPTVPGLSYY723 pKa = 10.56TIDD726 pKa = 3.35VRR728 pKa = 11.84QTDD731 pKa = 3.2AAGNVSTTVGSINFALAGAGSAPTVGLVNDD761 pKa = 3.98TGVVGDD767 pKa = 5.46AITSVGQLQITPAADD782 pKa = 3.26TAPNSTVQEE791 pKa = 4.3FSTDD795 pKa = 3.65GGQTWSTSFTPTSNAVNVVEE815 pKa = 4.32VRR817 pKa = 11.84YY818 pKa = 10.2NNPSDD823 pKa = 3.54TTGNSATLPASFTFTLDD840 pKa = 3.54TQVVAPTLSLQTGTSALTNSSAVTLGGIEE869 pKa = 4.09AGASVEE875 pKa = 4.0YY876 pKa = 9.8STDD879 pKa = 3.14GATWTAQQPVATQGSNTLYY898 pKa = 10.6VRR900 pKa = 11.84QTDD903 pKa = 3.63VAGNVSAASTALSFNLDD920 pKa = 3.44SVAPTAPVVVLTNGATTLTNNAALAALTGVEE951 pKa = 4.27AGALLEE957 pKa = 3.89YY958 pKa = 10.38RR959 pKa = 11.84VDD961 pKa = 3.46GGAWASSYY969 pKa = 7.77TAPTANAAHH978 pKa = 5.9TVDD981 pKa = 4.63VRR983 pKa = 11.84QTDD986 pKa = 3.18AAGNVSPTSSLSFTLDD1002 pKa = 3.01TLAPAALTPVLANGANGLTNSAALAALTPEE1032 pKa = 4.06VGALLEE1038 pKa = 3.96YY1039 pKa = 10.45RR1040 pKa = 11.84VDD1042 pKa = 3.46GGAWASSYY1050 pKa = 7.77TAPTANAAHH1059 pKa = 5.9TVDD1062 pKa = 4.63VRR1064 pKa = 11.84QTDD1067 pKa = 3.18AAGNVSPTSSLSFTLDD1083 pKa = 3.09TVAPAALGLALALANGANGLTNSAALAALTPEE1115 pKa = 4.06VGALLEE1121 pKa = 3.96YY1122 pKa = 10.45RR1123 pKa = 11.84VDD1125 pKa = 3.46GGAWASSYY1133 pKa = 7.77TAPTANAAHH1142 pKa = 5.9TVDD1145 pKa = 4.63VRR1147 pKa = 11.84QTDD1150 pKa = 3.24AAGNVSLASTPLAFTLDD1167 pKa = 3.42TVAPVALTVALLTDD1181 pKa = 3.77SADD1184 pKa = 3.32ATAAPAVFGAQIADD1198 pKa = 3.82KK1199 pKa = 9.75LTNDD1203 pKa = 3.26PTANFVPEE1211 pKa = 4.02TAGNLVEE1218 pKa = 4.21YY1219 pKa = 10.64SVDD1222 pKa = 5.42GINNWQPLSALAPGVLADD1240 pKa = 3.68GAYY1243 pKa = 10.58NLFVRR1248 pKa = 11.84EE1249 pKa = 3.44TDD1251 pKa = 3.05AAGNVTTSATGTYY1264 pKa = 10.39SFTLDD1269 pKa = 3.53TVSTAPTLALNAPILAGGVTYY1290 pKa = 10.66SQTGSFWDD1298 pKa = 3.78PLGGLLTGGTSTSTITDD1315 pKa = 3.41LHH1317 pKa = 7.03PSTEE1321 pKa = 4.3HH1322 pKa = 4.7YY1323 pKa = 9.43TYY1325 pKa = 11.18TDD1327 pKa = 3.2ATTGTPITTDD1337 pKa = 3.35LLGMQNYY1344 pKa = 10.2LLTLGGLINQTTGIGIPVSVTVTQTDD1370 pKa = 3.39AAGNASAPSAPLSFTFDD1387 pKa = 2.9NMTPLPVVGLATDD1400 pKa = 3.8SGRR1403 pKa = 11.84LNDD1406 pKa = 3.57VTTNQPTFSLLATEE1420 pKa = 4.6AQTTEE1425 pKa = 4.32AYY1427 pKa = 10.73SFDD1430 pKa = 3.14GMTWTSIPTLVNGTFTPTGLPIPAQVATPTAVPFWVQQTDD1470 pKa = 2.92ATGNVGTFMTTFMVDD1485 pKa = 3.15NFVAAPTVQATDD1497 pKa = 3.2STGALVGSGTLVSSGTLTLTGEE1519 pKa = 4.23TSLAPTYY1526 pKa = 10.24QVSTDD1531 pKa = 3.92SGITWVDD1538 pKa = 3.1SSIYY1542 pKa = 10.22TPTAMGANNILVHH1555 pKa = 6.55QIDD1558 pKa = 3.83DD1559 pKa = 4.05AGNISAATTFNLNLQGTVTNVTTGLSSAEE1588 pKa = 4.0AAGVSPFITLPTMSLTDD1605 pKa = 3.33SGIAGDD1611 pKa = 5.08HH1612 pKa = 5.61ITSNATVTLNAANLGGNVDD1631 pKa = 3.83TLSYY1635 pKa = 10.81SLNGGLWTAYY1645 pKa = 9.42TPGSSLPAVEE1655 pKa = 4.86GLNSLLVKK1663 pKa = 10.64EE1664 pKa = 5.0SVQSGAIGTLATHH1677 pKa = 6.4EE1678 pKa = 4.34TPIIPMSFTLDD1689 pKa = 3.47TVFPTAGATANNIAEE1704 pKa = 5.15LYY1706 pKa = 9.1TSASSGLSTAVVRR1719 pKa = 11.84VTYY1722 pKa = 10.78SEE1724 pKa = 4.18AVSVNAATLNFSGGGAAGGYY1744 pKa = 6.78TSQVVTSGASLDD1756 pKa = 3.95GAVSLGVQVDD1766 pKa = 4.12GSTVIEE1772 pKa = 4.06VRR1774 pKa = 11.84LAEE1777 pKa = 4.32TTLHH1781 pKa = 6.81AFTTAQVVPSVSLLITDD1798 pKa = 4.75LAGNQATVPIAVTNNPAPVFADD1820 pKa = 3.34WNLGMLPP1827 pKa = 3.87
MM1 pKa = 7.44NEE3 pKa = 3.95STVNPSVMPAIGPTQQGAGVNHH25 pKa = 7.43GGLSITPQRR34 pKa = 11.84HH35 pKa = 5.26EE36 pKa = 4.24GHH38 pKa = 6.37PTAAVTPAPLAHH50 pKa = 7.34RR51 pKa = 11.84FGQAPDD57 pKa = 3.68TEE59 pKa = 4.6GTEE62 pKa = 4.22EE63 pKa = 5.61DD64 pKa = 4.4SIATNAVEE72 pKa = 4.24TVEE75 pKa = 4.43TLDD78 pKa = 3.87EE79 pKa = 4.3DD80 pKa = 4.25TFGADD85 pKa = 3.06ISDD88 pKa = 3.63GTEE91 pKa = 3.75AFDD94 pKa = 4.5PGTMGHH100 pKa = 6.19EE101 pKa = 4.09SSGRR105 pKa = 11.84EE106 pKa = 3.56DD107 pKa = 3.48TLRR110 pKa = 11.84YY111 pKa = 9.37SALHH115 pKa = 6.35EE116 pKa = 4.2PAMMLTAADD125 pKa = 3.67ATFIASDD132 pKa = 3.95PAPLALAQPMEE143 pKa = 4.32YY144 pKa = 10.06LGSSAAASGGSDD156 pKa = 2.75AGAAAVGSTAMGEE169 pKa = 4.11LMPMLGAVTFGVIAATSANNATQAQTPNPSPTATGTAPPLVFSISHH215 pKa = 6.48AAGTPHH221 pKa = 7.49VMVVDD226 pKa = 3.8VTLPSTMVAGQTLTFSVAQGNAVLEE251 pKa = 4.4TVNYY255 pKa = 10.44VLTQTDD261 pKa = 3.47INAGSITKK269 pKa = 10.02FLGSNNLSDD278 pKa = 3.9SNYY281 pKa = 10.1QISVNQAGTTALASNTSTFHH301 pKa = 7.52LDD303 pKa = 2.98VTAPSAPQLNLSPNGNGTTVNVSLPIDD330 pKa = 3.72AVVGDD335 pKa = 4.68SLTLTLRR342 pKa = 11.84DD343 pKa = 3.57QTGSAVQTVTQAVTALNLSTHH364 pKa = 6.04TLQLSLNTGALPANTSYY381 pKa = 11.02QALANLTDD389 pKa = 3.22VSGNVSINSTPISMTTPGAPTPGITLTQDD418 pKa = 2.43SGTPGDD424 pKa = 4.73RR425 pKa = 11.84ITNQAGISLSGVLSSGVVEE444 pKa = 4.03YY445 pKa = 10.94SVDD448 pKa = 3.26GGAWTSVYY456 pKa = 8.83TAPTQNGNHH465 pKa = 5.3TVQVRR470 pKa = 11.84QFDD473 pKa = 4.28TNTGHH478 pKa = 6.32VSNPGSLSFTLDD490 pKa = 3.06TLAPAALIPVLANGANGLTNSAALAALTPEE520 pKa = 4.06VGALLEE526 pKa = 3.96YY527 pKa = 10.45RR528 pKa = 11.84VDD530 pKa = 3.46GGAWASSYY538 pKa = 7.73TAPTGDD544 pKa = 3.12IAHH547 pKa = 6.17TVDD550 pKa = 4.48VRR552 pKa = 11.84QTDD555 pKa = 3.27AAGNISAWFTLSFTLDD571 pKa = 3.24TVAPASVTPTLANGATTLTNNAALTALSATEE602 pKa = 3.78VGATVEE608 pKa = 3.97YY609 pKa = 10.42RR610 pKa = 11.84VDD612 pKa = 3.24GGAWGAYY619 pKa = 5.79TAPSAQGAHH628 pKa = 4.7TVEE631 pKa = 4.02ARR633 pKa = 11.84QVDD636 pKa = 3.65AAGNASAASAPLAFTLDD653 pKa = 3.39TQAPVAPGVALSNGNPAGSTTNNGSVSVTGIEE685 pKa = 3.84AGATVEE691 pKa = 4.06YY692 pKa = 10.23QINASGTWLPLVADD706 pKa = 3.84AQGAYY711 pKa = 9.52NVPSPTVPGLSYY723 pKa = 10.56TIDD726 pKa = 3.35VRR728 pKa = 11.84QTDD731 pKa = 3.2AAGNVSTTVGSINFALAGAGSAPTVGLVNDD761 pKa = 3.98TGVVGDD767 pKa = 5.46AITSVGQLQITPAADD782 pKa = 3.26TAPNSTVQEE791 pKa = 4.3FSTDD795 pKa = 3.65GGQTWSTSFTPTSNAVNVVEE815 pKa = 4.32VRR817 pKa = 11.84YY818 pKa = 10.2NNPSDD823 pKa = 3.54TTGNSATLPASFTFTLDD840 pKa = 3.54TQVVAPTLSLQTGTSALTNSSAVTLGGIEE869 pKa = 4.09AGASVEE875 pKa = 4.0YY876 pKa = 9.8STDD879 pKa = 3.14GATWTAQQPVATQGSNTLYY898 pKa = 10.6VRR900 pKa = 11.84QTDD903 pKa = 3.63VAGNVSAASTALSFNLDD920 pKa = 3.44SVAPTAPVVVLTNGATTLTNNAALAALTGVEE951 pKa = 4.27AGALLEE957 pKa = 3.89YY958 pKa = 10.38RR959 pKa = 11.84VDD961 pKa = 3.46GGAWASSYY969 pKa = 7.77TAPTANAAHH978 pKa = 5.9TVDD981 pKa = 4.63VRR983 pKa = 11.84QTDD986 pKa = 3.18AAGNVSPTSSLSFTLDD1002 pKa = 3.01TLAPAALTPVLANGANGLTNSAALAALTPEE1032 pKa = 4.06VGALLEE1038 pKa = 3.96YY1039 pKa = 10.45RR1040 pKa = 11.84VDD1042 pKa = 3.46GGAWASSYY1050 pKa = 7.77TAPTANAAHH1059 pKa = 5.9TVDD1062 pKa = 4.63VRR1064 pKa = 11.84QTDD1067 pKa = 3.18AAGNVSPTSSLSFTLDD1083 pKa = 3.09TVAPAALGLALALANGANGLTNSAALAALTPEE1115 pKa = 4.06VGALLEE1121 pKa = 3.96YY1122 pKa = 10.45RR1123 pKa = 11.84VDD1125 pKa = 3.46GGAWASSYY1133 pKa = 7.77TAPTANAAHH1142 pKa = 5.9TVDD1145 pKa = 4.63VRR1147 pKa = 11.84QTDD1150 pKa = 3.24AAGNVSLASTPLAFTLDD1167 pKa = 3.42TVAPVALTVALLTDD1181 pKa = 3.77SADD1184 pKa = 3.32ATAAPAVFGAQIADD1198 pKa = 3.82KK1199 pKa = 9.75LTNDD1203 pKa = 3.26PTANFVPEE1211 pKa = 4.02TAGNLVEE1218 pKa = 4.21YY1219 pKa = 10.64SVDD1222 pKa = 5.42GINNWQPLSALAPGVLADD1240 pKa = 3.68GAYY1243 pKa = 10.58NLFVRR1248 pKa = 11.84EE1249 pKa = 3.44TDD1251 pKa = 3.05AAGNVTTSATGTYY1264 pKa = 10.39SFTLDD1269 pKa = 3.53TVSTAPTLALNAPILAGGVTYY1290 pKa = 10.66SQTGSFWDD1298 pKa = 3.78PLGGLLTGGTSTSTITDD1315 pKa = 3.41LHH1317 pKa = 7.03PSTEE1321 pKa = 4.3HH1322 pKa = 4.7YY1323 pKa = 9.43TYY1325 pKa = 11.18TDD1327 pKa = 3.2ATTGTPITTDD1337 pKa = 3.35LLGMQNYY1344 pKa = 10.2LLTLGGLINQTTGIGIPVSVTVTQTDD1370 pKa = 3.39AAGNASAPSAPLSFTFDD1387 pKa = 2.9NMTPLPVVGLATDD1400 pKa = 3.8SGRR1403 pKa = 11.84LNDD1406 pKa = 3.57VTTNQPTFSLLATEE1420 pKa = 4.6AQTTEE1425 pKa = 4.32AYY1427 pKa = 10.73SFDD1430 pKa = 3.14GMTWTSIPTLVNGTFTPTGLPIPAQVATPTAVPFWVQQTDD1470 pKa = 2.92ATGNVGTFMTTFMVDD1485 pKa = 3.15NFVAAPTVQATDD1497 pKa = 3.2STGALVGSGTLVSSGTLTLTGEE1519 pKa = 4.23TSLAPTYY1526 pKa = 10.24QVSTDD1531 pKa = 3.92SGITWVDD1538 pKa = 3.1SSIYY1542 pKa = 10.22TPTAMGANNILVHH1555 pKa = 6.55QIDD1558 pKa = 3.83DD1559 pKa = 4.05AGNISAATTFNLNLQGTVTNVTTGLSSAEE1588 pKa = 4.0AAGVSPFITLPTMSLTDD1605 pKa = 3.33SGIAGDD1611 pKa = 5.08HH1612 pKa = 5.61ITSNATVTLNAANLGGNVDD1631 pKa = 3.83TLSYY1635 pKa = 10.81SLNGGLWTAYY1645 pKa = 9.42TPGSSLPAVEE1655 pKa = 4.86GLNSLLVKK1663 pKa = 10.64EE1664 pKa = 5.0SVQSGAIGTLATHH1677 pKa = 6.4EE1678 pKa = 4.34TPIIPMSFTLDD1689 pKa = 3.47TVFPTAGATANNIAEE1704 pKa = 5.15LYY1706 pKa = 9.1TSASSGLSTAVVRR1719 pKa = 11.84VTYY1722 pKa = 10.78SEE1724 pKa = 4.18AVSVNAATLNFSGGGAAGGYY1744 pKa = 6.78TSQVVTSGASLDD1756 pKa = 3.95GAVSLGVQVDD1766 pKa = 4.12GSTVIEE1772 pKa = 4.06VRR1774 pKa = 11.84LAEE1777 pKa = 4.32TTLHH1781 pKa = 6.81AFTTAQVVPSVSLLITDD1798 pKa = 4.75LAGNQATVPIAVTNNPAPVFADD1820 pKa = 3.34WNLGMLPP1827 pKa = 3.87
Molecular weight: 183.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4Z5K8|I4Z5K8_9BURK cAMP-binding protein OS=Leptothrix ochracea L12 OX=735332 GN=LepocDRAFT_00002310 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.05PLFHH6 pKa = 7.04VLLQFKK12 pKa = 9.95EE13 pKa = 4.27KK14 pKa = 10.7SRR16 pKa = 11.84LNTACCTAGCCVITSLHH33 pKa = 5.97RR34 pKa = 11.84RR35 pKa = 11.84PIRR38 pKa = 11.84RR39 pKa = 11.84MVLLSWINLQILRR52 pKa = 11.84SCHH55 pKa = 4.79QNRR58 pKa = 11.84VLTTNSNDD66 pKa = 3.53SCRR69 pKa = 11.84HH70 pKa = 5.42RR71 pKa = 11.84SNHH74 pKa = 4.87NNQRR78 pKa = 11.84PRR80 pKa = 11.84NHH82 pKa = 5.96QPNHH86 pKa = 6.58LYY88 pKa = 10.65LPPSKK93 pKa = 10.9GLFNTLFADD102 pKa = 3.94SSITGFFCILLIHH115 pKa = 7.22GYY117 pKa = 6.93TT118 pKa = 3.49
MM1 pKa = 7.5KK2 pKa = 10.05PLFHH6 pKa = 7.04VLLQFKK12 pKa = 9.95EE13 pKa = 4.27KK14 pKa = 10.7SRR16 pKa = 11.84LNTACCTAGCCVITSLHH33 pKa = 5.97RR34 pKa = 11.84RR35 pKa = 11.84PIRR38 pKa = 11.84RR39 pKa = 11.84MVLLSWINLQILRR52 pKa = 11.84SCHH55 pKa = 4.79QNRR58 pKa = 11.84VLTTNSNDD66 pKa = 3.53SCRR69 pKa = 11.84HH70 pKa = 5.42RR71 pKa = 11.84SNHH74 pKa = 4.87NNQRR78 pKa = 11.84PRR80 pKa = 11.84NHH82 pKa = 5.96QPNHH86 pKa = 6.58LYY88 pKa = 10.65LPPSKK93 pKa = 10.9GLFNTLFADD102 pKa = 3.94SSITGFFCILLIHH115 pKa = 7.22GYY117 pKa = 6.93TT118 pKa = 3.49
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
150642 |
20 |
1827 |
283.7 |
30.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.473 ± 0.14 | 1.069 ± 0.059 |
5.272 ± 0.074 | 5.006 ± 0.139 |
3.397 ± 0.074 | 7.956 ± 0.138 |
2.618 ± 0.073 | 4.703 ± 0.085 |
3.316 ± 0.116 | 10.7 ± 0.147 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.525 ± 0.068 | 2.861 ± 0.111 |
5.192 ± 0.078 | 4.257 ± 0.073 |
6.484 ± 0.164 | 6.045 ± 0.123 |
5.742 ± 0.163 | 7.586 ± 0.101 |
1.587 ± 0.054 | 2.212 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
