
Thalassotalea euphylliae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Colwelliaceae; Thalassotalea
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
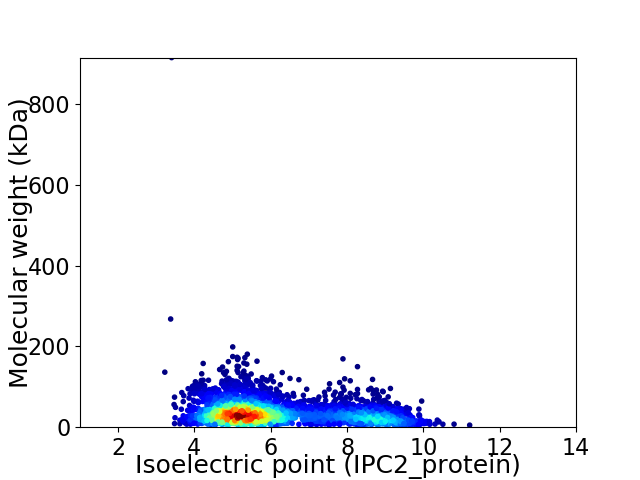
Virtual 2D-PAGE plot for 3642 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3E0U2V0|A0A3E0U2V0_9GAMM Antitoxin OS=Thalassotalea euphylliae OX=1655234 GN=DXX94_11645 PE=3 SV=1
MM1 pKa = 7.3KK2 pKa = 10.65NKK4 pKa = 8.42VTFPLNCIALILASQLTGCGSDD26 pKa = 4.14GDD28 pKa = 4.12NTPQIDD34 pKa = 5.33DD35 pKa = 4.0SATDD39 pKa = 3.44NTPPVITLIGNEE51 pKa = 4.45SIDD54 pKa = 3.59VVYY57 pKa = 10.98NAVFGDD63 pKa = 4.4LGAEE67 pKa = 4.03AHH69 pKa = 7.12DD70 pKa = 4.2EE71 pKa = 4.11TDD73 pKa = 3.12GAITVTSQGTIDD85 pKa = 3.25TAMPGTYY92 pKa = 9.89EE93 pKa = 3.92LTYY96 pKa = 9.98TATDD100 pKa = 3.24ASGNSSQATRR110 pKa = 11.84TVVVLEE116 pKa = 4.63DD117 pKa = 3.71TSQSATITTPQAPFSLFYY135 pKa = 11.27DD136 pKa = 4.54GVLKK140 pKa = 10.85GADD143 pKa = 3.48YY144 pKa = 10.12WSEE147 pKa = 3.93EE148 pKa = 3.95PQILSAGMGFDD159 pKa = 4.43GIIGIDD165 pKa = 3.61APEE168 pKa = 3.98LTFDD172 pKa = 3.96SVLEE176 pKa = 4.26AGGAWSSDD184 pKa = 3.38INCGTGAQNYY194 pKa = 7.16TSTSTLDD201 pKa = 3.58QIAGAYY207 pKa = 9.17INQTPYY213 pKa = 11.62GEE215 pKa = 4.76LKK217 pKa = 10.83DD218 pKa = 3.91GAIGLDD224 pKa = 3.42GLPIVLSWPIDD235 pKa = 3.52TSTLSLTDD243 pKa = 3.67FQFTLNTGDD252 pKa = 3.13IVRR255 pKa = 11.84PLAIGPLPNFEE266 pKa = 4.76DD267 pKa = 3.88NEE269 pKa = 4.51RR270 pKa = 11.84NTPVAFGEE278 pKa = 4.24FGNRR282 pKa = 11.84LPSDD286 pKa = 3.65HH287 pKa = 7.63PDD289 pKa = 2.61ARR291 pKa = 11.84FPIKK295 pKa = 10.78LEE297 pKa = 3.79IVEE300 pKa = 5.24DD301 pKa = 4.07DD302 pKa = 4.1TPLMMVGPGGQVVSAVGLTWEE323 pKa = 4.79TDD325 pKa = 3.06SSPYY329 pKa = 10.52DD330 pKa = 3.71EE331 pKa = 5.54NNGPRR336 pKa = 11.84LVGAKK341 pKa = 9.94LNRR344 pKa = 11.84IEE346 pKa = 4.09GQMDD350 pKa = 3.9GEE352 pKa = 5.08GISTPIPLIPANDD365 pKa = 3.37ATVLYY370 pKa = 10.79DD371 pKa = 3.94EE372 pKa = 5.2GDD374 pKa = 3.17FMLRR378 pKa = 11.84MLTTGGFSPDD388 pKa = 3.4GVSGLTPNDD397 pKa = 3.17FEE399 pKa = 5.16RR400 pKa = 11.84FFRR403 pKa = 11.84IHH405 pKa = 6.9ANGVDD410 pKa = 3.69GSTVIIDD417 pKa = 3.81KK418 pKa = 10.97VGEE421 pKa = 4.07EE422 pKa = 4.44YY423 pKa = 10.77AVQGGTLRR431 pKa = 11.84VVGMSDD437 pKa = 3.35LGQPEE442 pKa = 4.15GGEE445 pKa = 4.42VVFDD449 pKa = 3.64ACYY452 pKa = 10.83SEE454 pKa = 6.09DD455 pKa = 3.62RR456 pKa = 11.84DD457 pKa = 3.83NYY459 pKa = 10.24IDD461 pKa = 4.11IIMVGDD467 pKa = 3.75EE468 pKa = 3.66EE469 pKa = 4.19AARR472 pKa = 11.84NITILEE478 pKa = 4.16IPSLEE483 pKa = 4.03EE484 pKa = 4.56GYY486 pKa = 10.55SAFYY490 pKa = 10.93NPGGPGSTPFEE501 pKa = 4.29GVSYY505 pKa = 7.74TAPGPRR511 pKa = 11.84DD512 pKa = 3.86LEE514 pKa = 4.16PVIIALDD521 pKa = 3.5DD522 pKa = 3.99PMRR525 pKa = 11.84VTYY528 pKa = 10.7VPGSEE533 pKa = 4.37NVEE536 pKa = 4.26TPDD539 pKa = 3.9NLMTFLFEE547 pKa = 4.75GIEE550 pKa = 4.12RR551 pKa = 11.84EE552 pKa = 4.1YY553 pKa = 10.75LLNMSSNYY561 pKa = 9.98TGDD564 pKa = 3.53TPVPLLFDD572 pKa = 3.37FHH574 pKa = 7.51GLNGTAEE581 pKa = 4.11QQNTDD586 pKa = 3.08SQFNQLAEE594 pKa = 4.24TEE596 pKa = 4.16NFILVTPKK604 pKa = 10.76AIGGWNVTGFPLGGDD619 pKa = 3.52ANDD622 pKa = 4.62LGFIDD627 pKa = 5.05ALINEE632 pKa = 5.25LSTTYY637 pKa = 10.84NIDD640 pKa = 3.22TNRR643 pKa = 11.84IYY645 pKa = 10.99AAGFSLGGFFSFEE658 pKa = 4.08LACQYY663 pKa = 11.38SDD665 pKa = 3.16TFAAIAPVSGVMTPNMANDD684 pKa = 4.38CLPEE688 pKa = 4.68RR689 pKa = 11.84PIPILQTHH697 pKa = 5.99GTADD701 pKa = 3.56DD702 pKa = 3.78QLPYY706 pKa = 10.92AQAQTVLQWWIDD718 pKa = 3.57FNQTDD723 pKa = 4.37VEE725 pKa = 4.13PAITDD730 pKa = 3.97LEE732 pKa = 4.45DD733 pKa = 3.38TFPDD737 pKa = 3.15NGTTVQRR744 pKa = 11.84YY745 pKa = 8.35VYY747 pKa = 10.86GNGTNGVSVEE757 pKa = 3.91HH758 pKa = 6.3LRR760 pKa = 11.84IEE762 pKa = 4.74GGQHH766 pKa = 5.6IWPGSAGDD774 pKa = 3.78SDD776 pKa = 4.21INMAEE781 pKa = 4.41EE782 pKa = 3.58IWSFFSNYY790 pKa = 10.0DD791 pKa = 3.75LNGKK795 pKa = 9.15IPDD798 pKa = 3.48
MM1 pKa = 7.3KK2 pKa = 10.65NKK4 pKa = 8.42VTFPLNCIALILASQLTGCGSDD26 pKa = 4.14GDD28 pKa = 4.12NTPQIDD34 pKa = 5.33DD35 pKa = 4.0SATDD39 pKa = 3.44NTPPVITLIGNEE51 pKa = 4.45SIDD54 pKa = 3.59VVYY57 pKa = 10.98NAVFGDD63 pKa = 4.4LGAEE67 pKa = 4.03AHH69 pKa = 7.12DD70 pKa = 4.2EE71 pKa = 4.11TDD73 pKa = 3.12GAITVTSQGTIDD85 pKa = 3.25TAMPGTYY92 pKa = 9.89EE93 pKa = 3.92LTYY96 pKa = 9.98TATDD100 pKa = 3.24ASGNSSQATRR110 pKa = 11.84TVVVLEE116 pKa = 4.63DD117 pKa = 3.71TSQSATITTPQAPFSLFYY135 pKa = 11.27DD136 pKa = 4.54GVLKK140 pKa = 10.85GADD143 pKa = 3.48YY144 pKa = 10.12WSEE147 pKa = 3.93EE148 pKa = 3.95PQILSAGMGFDD159 pKa = 4.43GIIGIDD165 pKa = 3.61APEE168 pKa = 3.98LTFDD172 pKa = 3.96SVLEE176 pKa = 4.26AGGAWSSDD184 pKa = 3.38INCGTGAQNYY194 pKa = 7.16TSTSTLDD201 pKa = 3.58QIAGAYY207 pKa = 9.17INQTPYY213 pKa = 11.62GEE215 pKa = 4.76LKK217 pKa = 10.83DD218 pKa = 3.91GAIGLDD224 pKa = 3.42GLPIVLSWPIDD235 pKa = 3.52TSTLSLTDD243 pKa = 3.67FQFTLNTGDD252 pKa = 3.13IVRR255 pKa = 11.84PLAIGPLPNFEE266 pKa = 4.76DD267 pKa = 3.88NEE269 pKa = 4.51RR270 pKa = 11.84NTPVAFGEE278 pKa = 4.24FGNRR282 pKa = 11.84LPSDD286 pKa = 3.65HH287 pKa = 7.63PDD289 pKa = 2.61ARR291 pKa = 11.84FPIKK295 pKa = 10.78LEE297 pKa = 3.79IVEE300 pKa = 5.24DD301 pKa = 4.07DD302 pKa = 4.1TPLMMVGPGGQVVSAVGLTWEE323 pKa = 4.79TDD325 pKa = 3.06SSPYY329 pKa = 10.52DD330 pKa = 3.71EE331 pKa = 5.54NNGPRR336 pKa = 11.84LVGAKK341 pKa = 9.94LNRR344 pKa = 11.84IEE346 pKa = 4.09GQMDD350 pKa = 3.9GEE352 pKa = 5.08GISTPIPLIPANDD365 pKa = 3.37ATVLYY370 pKa = 10.79DD371 pKa = 3.94EE372 pKa = 5.2GDD374 pKa = 3.17FMLRR378 pKa = 11.84MLTTGGFSPDD388 pKa = 3.4GVSGLTPNDD397 pKa = 3.17FEE399 pKa = 5.16RR400 pKa = 11.84FFRR403 pKa = 11.84IHH405 pKa = 6.9ANGVDD410 pKa = 3.69GSTVIIDD417 pKa = 3.81KK418 pKa = 10.97VGEE421 pKa = 4.07EE422 pKa = 4.44YY423 pKa = 10.77AVQGGTLRR431 pKa = 11.84VVGMSDD437 pKa = 3.35LGQPEE442 pKa = 4.15GGEE445 pKa = 4.42VVFDD449 pKa = 3.64ACYY452 pKa = 10.83SEE454 pKa = 6.09DD455 pKa = 3.62RR456 pKa = 11.84DD457 pKa = 3.83NYY459 pKa = 10.24IDD461 pKa = 4.11IIMVGDD467 pKa = 3.75EE468 pKa = 3.66EE469 pKa = 4.19AARR472 pKa = 11.84NITILEE478 pKa = 4.16IPSLEE483 pKa = 4.03EE484 pKa = 4.56GYY486 pKa = 10.55SAFYY490 pKa = 10.93NPGGPGSTPFEE501 pKa = 4.29GVSYY505 pKa = 7.74TAPGPRR511 pKa = 11.84DD512 pKa = 3.86LEE514 pKa = 4.16PVIIALDD521 pKa = 3.5DD522 pKa = 3.99PMRR525 pKa = 11.84VTYY528 pKa = 10.7VPGSEE533 pKa = 4.37NVEE536 pKa = 4.26TPDD539 pKa = 3.9NLMTFLFEE547 pKa = 4.75GIEE550 pKa = 4.12RR551 pKa = 11.84EE552 pKa = 4.1YY553 pKa = 10.75LLNMSSNYY561 pKa = 9.98TGDD564 pKa = 3.53TPVPLLFDD572 pKa = 3.37FHH574 pKa = 7.51GLNGTAEE581 pKa = 4.11QQNTDD586 pKa = 3.08SQFNQLAEE594 pKa = 4.24TEE596 pKa = 4.16NFILVTPKK604 pKa = 10.76AIGGWNVTGFPLGGDD619 pKa = 3.52ANDD622 pKa = 4.62LGFIDD627 pKa = 5.05ALINEE632 pKa = 5.25LSTTYY637 pKa = 10.84NIDD640 pKa = 3.22TNRR643 pKa = 11.84IYY645 pKa = 10.99AAGFSLGGFFSFEE658 pKa = 4.08LACQYY663 pKa = 11.38SDD665 pKa = 3.16TFAAIAPVSGVMTPNMANDD684 pKa = 4.38CLPEE688 pKa = 4.68RR689 pKa = 11.84PIPILQTHH697 pKa = 5.99GTADD701 pKa = 3.56DD702 pKa = 3.78QLPYY706 pKa = 10.92AQAQTVLQWWIDD718 pKa = 3.57FNQTDD723 pKa = 4.37VEE725 pKa = 4.13PAITDD730 pKa = 3.97LEE732 pKa = 4.45DD733 pKa = 3.38TFPDD737 pKa = 3.15NGTTVQRR744 pKa = 11.84YY745 pKa = 8.35VYY747 pKa = 10.86GNGTNGVSVEE757 pKa = 3.91HH758 pKa = 6.3LRR760 pKa = 11.84IEE762 pKa = 4.74GGQHH766 pKa = 5.6IWPGSAGDD774 pKa = 3.78SDD776 pKa = 4.21INMAEE781 pKa = 4.41EE782 pKa = 3.58IWSFFSNYY790 pKa = 10.0DD791 pKa = 3.75LNGKK795 pKa = 9.15IPDD798 pKa = 3.48
Molecular weight: 85.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3E0TII7|A0A3E0TII7_9GAMM Carboxy-S-adenosyl-L-methionine synthase OS=Thalassotalea euphylliae OX=1655234 GN=cmoA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.49GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.49GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1233835 |
38 |
8735 |
338.8 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.219 ± 0.039 | 0.927 ± 0.014 |
5.566 ± 0.042 | 6.145 ± 0.039 |
4.199 ± 0.027 | 6.506 ± 0.052 |
2.211 ± 0.021 | 6.364 ± 0.029 |
5.347 ± 0.053 | 10.234 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.332 ± 0.024 | 4.636 ± 0.038 |
3.719 ± 0.025 | 5.166 ± 0.04 |
4.143 ± 0.032 | 6.727 ± 0.04 |
5.464 ± 0.04 | 6.9 ± 0.031 |
1.142 ± 0.014 | 3.056 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
