
Roseisalinus antarcticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseisalinus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
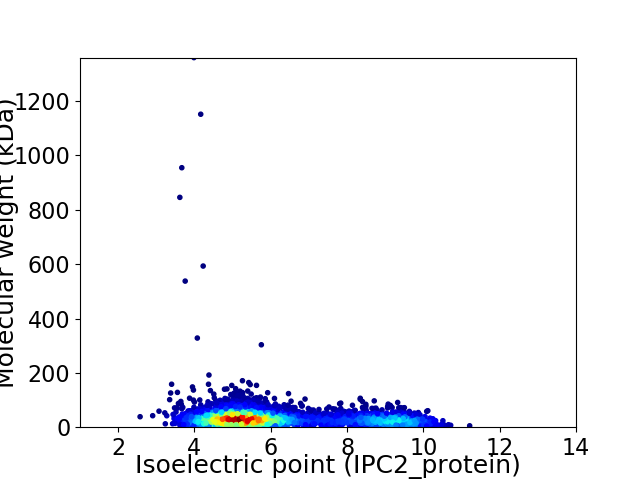
Virtual 2D-PAGE plot for 4698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y5SNP9|A0A1Y5SNP9_9RHOB Pyridoxine 5'-phosphate synthase OS=Roseisalinus antarcticus OX=254357 GN=pdxJ PE=3 SV=1
MM1 pKa = 7.54TLRR4 pKa = 11.84ISLILGPVAACALALPAAAQDD25 pKa = 3.92YY26 pKa = 10.53CGGGSGTWIGGNQANSDD43 pKa = 3.38IATSDD48 pKa = 3.38TYY50 pKa = 11.02RR51 pKa = 11.84EE52 pKa = 3.84QLALVLLGNAHH63 pKa = 6.31VSLFDD68 pKa = 4.25LSSAMDD74 pKa = 3.45VRR76 pKa = 11.84LEE78 pKa = 3.97AQGRR82 pKa = 11.84GAGDD86 pKa = 3.53PVIEE90 pKa = 4.67ILDD93 pKa = 3.61STGAVVGGDD102 pKa = 3.74DD103 pKa = 4.53DD104 pKa = 5.24SGGGTSSAALLSLQPGTYY122 pKa = 10.59CMNTSSYY129 pKa = 11.87DD130 pKa = 3.79NAPLTATVRR139 pKa = 11.84IGQTRR144 pKa = 11.84HH145 pKa = 4.89EE146 pKa = 4.43ALTEE150 pKa = 4.21GGAQLSANGGNTGGGNTGGGDD171 pKa = 3.34TGNDD175 pKa = 3.63GGGSMTDD182 pKa = 3.06SGACGGAMMIGGGNPVEE199 pKa = 4.47GSLGGGLSVTNAIDD213 pKa = 3.84SVPTYY218 pKa = 10.51AFVVGSPTTLSITAEE233 pKa = 4.11NQDD236 pKa = 3.82ADD238 pKa = 3.99PVLTLTGDD246 pKa = 3.57DD247 pKa = 3.51GTFYY251 pKa = 11.4AEE253 pKa = 4.7NDD255 pKa = 3.88DD256 pKa = 4.81FDD258 pKa = 4.6GLNSRR263 pKa = 11.84IDD265 pKa = 3.54MTTALQPGTYY275 pKa = 9.86CIGLRR280 pKa = 11.84ALSNGTLPVTVALTGYY296 pKa = 10.4DD297 pKa = 3.55PVAAAQALYY306 pKa = 10.54DD307 pKa = 3.89QGEE310 pKa = 4.44VAPPMDD316 pKa = 3.98GSYY319 pKa = 10.67PITPLGTVASRR330 pKa = 11.84SVSNAQVQGDD340 pKa = 3.76RR341 pKa = 11.84AQWFSLTVDD350 pKa = 3.2QTGLLLIEE358 pKa = 5.92AIATSNGDD366 pKa = 3.28PVLRR370 pKa = 11.84LFDD373 pKa = 3.97DD374 pKa = 3.75VGRR377 pKa = 11.84QIGYY381 pKa = 10.04NDD383 pKa = 5.17DD384 pKa = 3.66YY385 pKa = 11.86GQGLDD390 pKa = 3.37SQIAARR396 pKa = 11.84VFPGTYY402 pKa = 10.02LVALTDD408 pKa = 3.87ISGSSPMLRR417 pKa = 11.84LVMEE421 pKa = 5.05RR422 pKa = 11.84YY423 pKa = 9.83VPAQQ427 pKa = 3.09
MM1 pKa = 7.54TLRR4 pKa = 11.84ISLILGPVAACALALPAAAQDD25 pKa = 3.92YY26 pKa = 10.53CGGGSGTWIGGNQANSDD43 pKa = 3.38IATSDD48 pKa = 3.38TYY50 pKa = 11.02RR51 pKa = 11.84EE52 pKa = 3.84QLALVLLGNAHH63 pKa = 6.31VSLFDD68 pKa = 4.25LSSAMDD74 pKa = 3.45VRR76 pKa = 11.84LEE78 pKa = 3.97AQGRR82 pKa = 11.84GAGDD86 pKa = 3.53PVIEE90 pKa = 4.67ILDD93 pKa = 3.61STGAVVGGDD102 pKa = 3.74DD103 pKa = 4.53DD104 pKa = 5.24SGGGTSSAALLSLQPGTYY122 pKa = 10.59CMNTSSYY129 pKa = 11.87DD130 pKa = 3.79NAPLTATVRR139 pKa = 11.84IGQTRR144 pKa = 11.84HH145 pKa = 4.89EE146 pKa = 4.43ALTEE150 pKa = 4.21GGAQLSANGGNTGGGNTGGGDD171 pKa = 3.34TGNDD175 pKa = 3.63GGGSMTDD182 pKa = 3.06SGACGGAMMIGGGNPVEE199 pKa = 4.47GSLGGGLSVTNAIDD213 pKa = 3.84SVPTYY218 pKa = 10.51AFVVGSPTTLSITAEE233 pKa = 4.11NQDD236 pKa = 3.82ADD238 pKa = 3.99PVLTLTGDD246 pKa = 3.57DD247 pKa = 3.51GTFYY251 pKa = 11.4AEE253 pKa = 4.7NDD255 pKa = 3.88DD256 pKa = 4.81FDD258 pKa = 4.6GLNSRR263 pKa = 11.84IDD265 pKa = 3.54MTTALQPGTYY275 pKa = 9.86CIGLRR280 pKa = 11.84ALSNGTLPVTVALTGYY296 pKa = 10.4DD297 pKa = 3.55PVAAAQALYY306 pKa = 10.54DD307 pKa = 3.89QGEE310 pKa = 4.44VAPPMDD316 pKa = 3.98GSYY319 pKa = 10.67PITPLGTVASRR330 pKa = 11.84SVSNAQVQGDD340 pKa = 3.76RR341 pKa = 11.84AQWFSLTVDD350 pKa = 3.2QTGLLLIEE358 pKa = 5.92AIATSNGDD366 pKa = 3.28PVLRR370 pKa = 11.84LFDD373 pKa = 3.97DD374 pKa = 3.75VGRR377 pKa = 11.84QIGYY381 pKa = 10.04NDD383 pKa = 5.17DD384 pKa = 3.66YY385 pKa = 11.86GQGLDD390 pKa = 3.37SQIAARR396 pKa = 11.84VFPGTYY402 pKa = 10.02LVALTDD408 pKa = 3.87ISGSSPMLRR417 pKa = 11.84LVMEE421 pKa = 5.05RR422 pKa = 11.84YY423 pKa = 9.83VPAQQ427 pKa = 3.09
Molecular weight: 43.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y5TNX7|A0A1Y5TNX7_9RHOB Putative FAD-linked oxidoreductase OS=Roseisalinus antarcticus OX=254357 GN=ROA7023_03318 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84KK29 pKa = 8.8ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84KK29 pKa = 8.8ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1473940 |
29 |
12848 |
313.7 |
33.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.838 ± 0.06 | 0.839 ± 0.014 |
6.332 ± 0.062 | 5.793 ± 0.037 |
3.579 ± 0.024 | 9.189 ± 0.055 |
1.959 ± 0.024 | 4.916 ± 0.024 |
2.376 ± 0.037 | 10.016 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.588 ± 0.031 | 2.295 ± 0.021 |
5.34 ± 0.036 | 2.884 ± 0.02 |
7.339 ± 0.047 | 5.118 ± 0.032 |
5.672 ± 0.048 | 7.382 ± 0.034 |
1.454 ± 0.017 | 2.093 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
