
Halomicronema hongdechloris C2206
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Prochlorotrichaceae; Halomicronema; Halomicronema hongdechloris
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
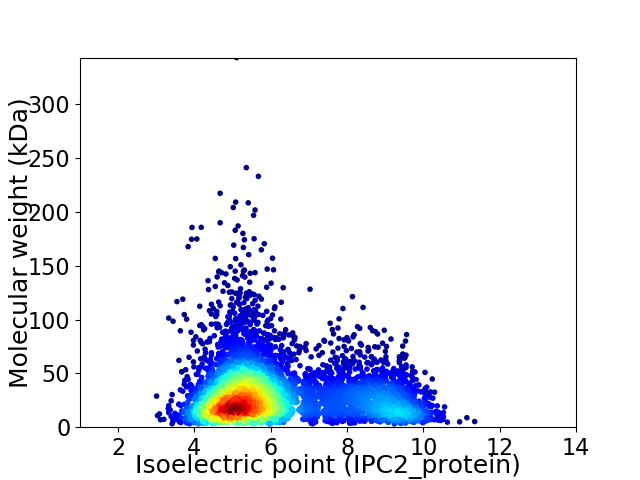
Virtual 2D-PAGE plot for 5211 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z3HUC4|A0A1Z3HUC4_9CYAN Tetratricopeptide repeat domain protein OS=Halomicronema hongdechloris C2206 OX=1641165 GN=XM38_048670 PE=4 SV=1
MM1 pKa = 7.76SKK3 pKa = 10.72LLWKK7 pKa = 10.53SLLAAPAALGAVLAISGTSVAAEE30 pKa = 3.81SVEE33 pKa = 3.89PSFNEE38 pKa = 3.81LSAAEE43 pKa = 4.22PAQLAQVTSVDD54 pKa = 3.76EE55 pKa = 4.57LSDD58 pKa = 3.72VLPSDD63 pKa = 3.25WAYY66 pKa = 9.08TALQNLVEE74 pKa = 4.45TYY76 pKa = 10.83GCIEE80 pKa = 4.99GYY82 pKa = 9.35PNRR85 pKa = 11.84TFRR88 pKa = 11.84GDD90 pKa = 3.28RR91 pKa = 11.84ALTRR95 pKa = 11.84YY96 pKa = 8.99EE97 pKa = 3.9FAAGLNACLDD107 pKa = 4.09VISNLIVGGGVGEE120 pKa = 4.48SDD122 pKa = 3.47LATIRR127 pKa = 11.84RR128 pKa = 11.84LQEE131 pKa = 3.45EE132 pKa = 4.51FQAEE136 pKa = 4.36LATLRR141 pKa = 11.84GRR143 pKa = 11.84VDD145 pKa = 3.18ALEE148 pKa = 4.62ADD150 pKa = 4.06VAEE153 pKa = 4.67LEE155 pKa = 4.39ANQFSTTTKK164 pKa = 10.51LRR166 pKa = 11.84GQVDD170 pKa = 3.42SHH172 pKa = 7.2IVAPFDD178 pKa = 3.53EE179 pKa = 5.08LEE181 pKa = 4.21GVEE184 pKa = 5.52DD185 pKa = 3.57STTFTNRR192 pKa = 11.84VRR194 pKa = 11.84MNFDD198 pKa = 3.05TSFTGEE204 pKa = 3.53DD205 pKa = 3.22RR206 pKa = 11.84LRR208 pKa = 11.84VRR210 pKa = 11.84LQASGGADD218 pKa = 3.27PLVAGGGLANGDD230 pKa = 4.11DD231 pKa = 3.81ASGDD235 pKa = 2.95GDD237 pKa = 4.44YY238 pKa = 11.47NVDD241 pKa = 3.48IDD243 pKa = 3.97DD244 pKa = 4.87FYY246 pKa = 12.0YY247 pKa = 10.88LFPVGDD253 pKa = 3.89RR254 pKa = 11.84LDD256 pKa = 4.0IIIAANGIVTDD267 pKa = 4.63DD268 pKa = 4.0YY269 pKa = 11.55VVSTIVPFDD278 pKa = 4.0GPSVADD284 pKa = 3.48PGGPVFYY291 pKa = 10.35DD292 pKa = 3.59FDD294 pKa = 3.96MGGSAGAGFSFALTNNIAIDD314 pKa = 3.37AGYY317 pKa = 10.73SFDD320 pKa = 3.91EE321 pKa = 5.12AEE323 pKa = 4.63GADD326 pKa = 3.86PLIGISAASEE336 pKa = 3.6QSYY339 pKa = 10.15IGQVTFISDD348 pKa = 4.84GILDD352 pKa = 3.89LAGTFIRR359 pKa = 11.84GDD361 pKa = 3.59SGDD364 pKa = 3.74GAFTNTFAALANLDD378 pKa = 3.77FGRR381 pKa = 11.84FMIGGYY387 pKa = 10.1FSYY390 pKa = 10.8HH391 pKa = 6.87DD392 pKa = 4.8LDD394 pKa = 3.77GTPAVGDD401 pKa = 4.04DD402 pKa = 4.69DD403 pKa = 5.8FTTSWQAGIAVPDD416 pKa = 3.98LFIEE420 pKa = 5.37GAQLGAYY427 pKa = 7.36YY428 pKa = 9.55TALPEE433 pKa = 4.18YY434 pKa = 10.56ASGVNPYY441 pKa = 9.22MIEE444 pKa = 4.23GYY446 pKa = 10.73YY447 pKa = 10.12SIPVNQFLTITPALIYY463 pKa = 10.96GDD465 pKa = 4.17IDD467 pKa = 3.45SGAADD472 pKa = 3.42EE473 pKa = 4.29EE474 pKa = 4.48AFYY477 pKa = 10.98GAIRR481 pKa = 11.84ATFEE485 pKa = 3.85FF486 pKa = 4.77
MM1 pKa = 7.76SKK3 pKa = 10.72LLWKK7 pKa = 10.53SLLAAPAALGAVLAISGTSVAAEE30 pKa = 3.81SVEE33 pKa = 3.89PSFNEE38 pKa = 3.81LSAAEE43 pKa = 4.22PAQLAQVTSVDD54 pKa = 3.76EE55 pKa = 4.57LSDD58 pKa = 3.72VLPSDD63 pKa = 3.25WAYY66 pKa = 9.08TALQNLVEE74 pKa = 4.45TYY76 pKa = 10.83GCIEE80 pKa = 4.99GYY82 pKa = 9.35PNRR85 pKa = 11.84TFRR88 pKa = 11.84GDD90 pKa = 3.28RR91 pKa = 11.84ALTRR95 pKa = 11.84YY96 pKa = 8.99EE97 pKa = 3.9FAAGLNACLDD107 pKa = 4.09VISNLIVGGGVGEE120 pKa = 4.48SDD122 pKa = 3.47LATIRR127 pKa = 11.84RR128 pKa = 11.84LQEE131 pKa = 3.45EE132 pKa = 4.51FQAEE136 pKa = 4.36LATLRR141 pKa = 11.84GRR143 pKa = 11.84VDD145 pKa = 3.18ALEE148 pKa = 4.62ADD150 pKa = 4.06VAEE153 pKa = 4.67LEE155 pKa = 4.39ANQFSTTTKK164 pKa = 10.51LRR166 pKa = 11.84GQVDD170 pKa = 3.42SHH172 pKa = 7.2IVAPFDD178 pKa = 3.53EE179 pKa = 5.08LEE181 pKa = 4.21GVEE184 pKa = 5.52DD185 pKa = 3.57STTFTNRR192 pKa = 11.84VRR194 pKa = 11.84MNFDD198 pKa = 3.05TSFTGEE204 pKa = 3.53DD205 pKa = 3.22RR206 pKa = 11.84LRR208 pKa = 11.84VRR210 pKa = 11.84LQASGGADD218 pKa = 3.27PLVAGGGLANGDD230 pKa = 4.11DD231 pKa = 3.81ASGDD235 pKa = 2.95GDD237 pKa = 4.44YY238 pKa = 11.47NVDD241 pKa = 3.48IDD243 pKa = 3.97DD244 pKa = 4.87FYY246 pKa = 12.0YY247 pKa = 10.88LFPVGDD253 pKa = 3.89RR254 pKa = 11.84LDD256 pKa = 4.0IIIAANGIVTDD267 pKa = 4.63DD268 pKa = 4.0YY269 pKa = 11.55VVSTIVPFDD278 pKa = 4.0GPSVADD284 pKa = 3.48PGGPVFYY291 pKa = 10.35DD292 pKa = 3.59FDD294 pKa = 3.96MGGSAGAGFSFALTNNIAIDD314 pKa = 3.37AGYY317 pKa = 10.73SFDD320 pKa = 3.91EE321 pKa = 5.12AEE323 pKa = 4.63GADD326 pKa = 3.86PLIGISAASEE336 pKa = 3.6QSYY339 pKa = 10.15IGQVTFISDD348 pKa = 4.84GILDD352 pKa = 3.89LAGTFIRR359 pKa = 11.84GDD361 pKa = 3.59SGDD364 pKa = 3.74GAFTNTFAALANLDD378 pKa = 3.77FGRR381 pKa = 11.84FMIGGYY387 pKa = 10.1FSYY390 pKa = 10.8HH391 pKa = 6.87DD392 pKa = 4.8LDD394 pKa = 3.77GTPAVGDD401 pKa = 4.04DD402 pKa = 4.69DD403 pKa = 5.8FTTSWQAGIAVPDD416 pKa = 3.98LFIEE420 pKa = 5.37GAQLGAYY427 pKa = 7.36YY428 pKa = 9.55TALPEE433 pKa = 4.18YY434 pKa = 10.56ASGVNPYY441 pKa = 9.22MIEE444 pKa = 4.23GYY446 pKa = 10.73YY447 pKa = 10.12SIPVNQFLTITPALIYY463 pKa = 10.96GDD465 pKa = 4.17IDD467 pKa = 3.45SGAADD472 pKa = 3.42EE473 pKa = 4.29EE474 pKa = 4.48AFYY477 pKa = 10.98GAIRR481 pKa = 11.84ATFEE485 pKa = 3.85FF486 pKa = 4.77
Molecular weight: 51.48 kDa
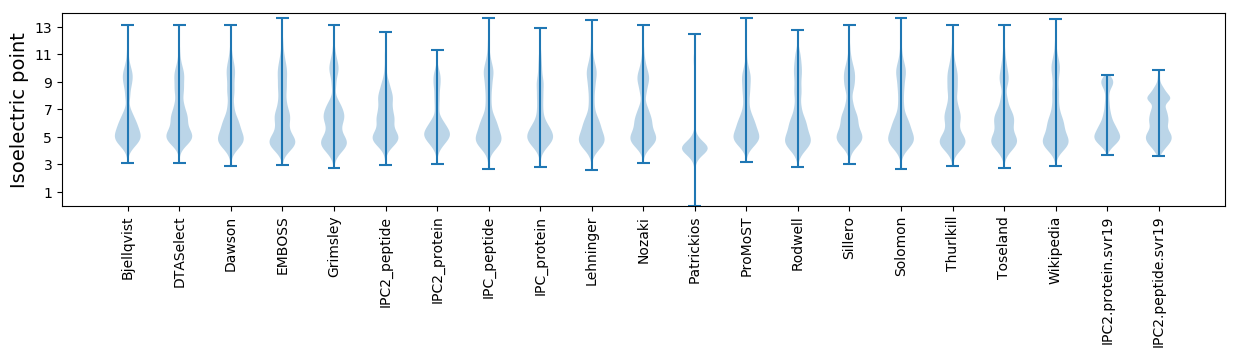
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z3HGK3|A0A1Z3HGK3_9CYAN Uncharacterized protein OS=Halomicronema hongdechloris C2206 OX=1641165 GN=XM38_003020 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 8.94RR4 pKa = 11.84TLGGTVRR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.12RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TQTGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.54GRR40 pKa = 11.84ARR42 pKa = 11.84LAVV45 pKa = 3.42
MM1 pKa = 7.55TKK3 pKa = 8.94RR4 pKa = 11.84TLGGTVRR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.12RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TQTGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.54GRR40 pKa = 11.84ARR42 pKa = 11.84LAVV45 pKa = 3.42
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1532136 |
29 |
3109 |
294.0 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.726 ± 0.038 | 1.019 ± 0.011 |
5.417 ± 0.03 | 5.6 ± 0.036 |
3.489 ± 0.021 | 7.101 ± 0.035 |
2.292 ± 0.02 | 5.296 ± 0.027 |
2.784 ± 0.029 | 11.66 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.954 ± 0.017 | 2.91 ± 0.023 |
5.553 ± 0.03 | 5.946 ± 0.036 |
6.375 ± 0.03 | 5.967 ± 0.026 |
5.626 ± 0.023 | 6.738 ± 0.028 |
1.679 ± 0.016 | 2.866 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
