
Alteromonadaceae bacterium Bs31
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; unclassified Alteromonadaceae
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
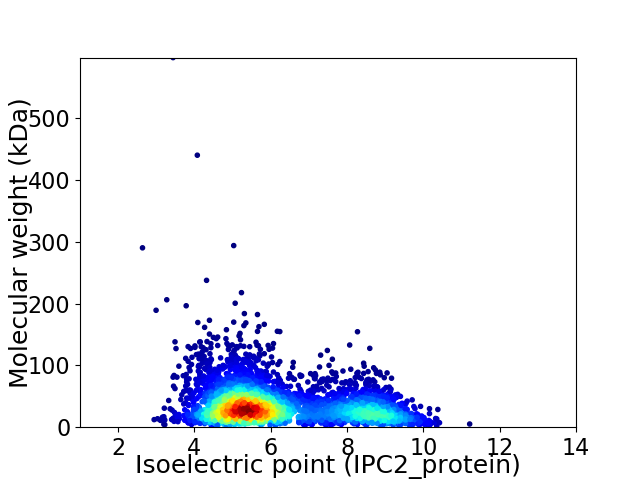
Virtual 2D-PAGE plot for 4297 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
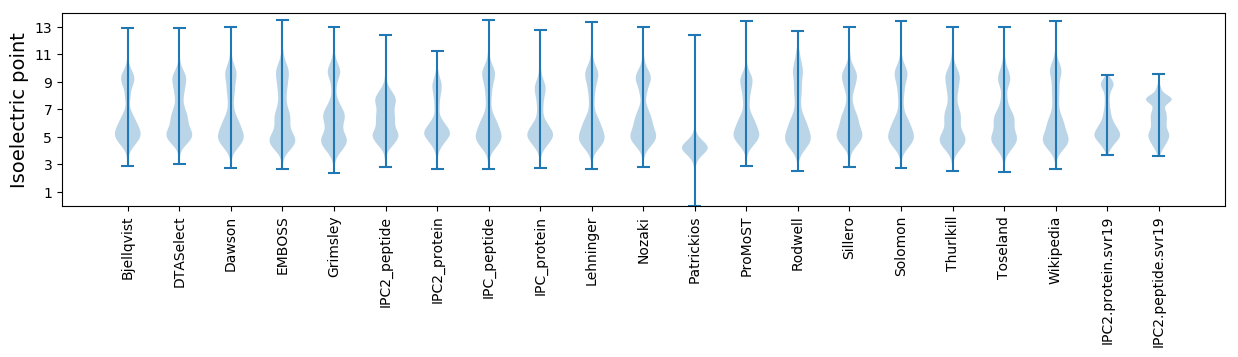
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y6CG80|A0A1Y6CG80_9ALTE DNA topoisomerase 4 subunit B OS=Alteromonadaceae bacterium Bs31 OX=1304903 GN=parE PE=3 SV=1
MM1 pKa = 7.53KK2 pKa = 9.98PLAQKK7 pKa = 10.04IHH9 pKa = 6.45LALLTALLLGGCGGDD24 pKa = 3.38SDD26 pKa = 5.51SPGGDD31 pKa = 2.74EE32 pKa = 5.72SGGNTNLQQNNNMTGVINPVTDD54 pKa = 3.44VDD56 pKa = 3.72WYY58 pKa = 10.6SIQVNEE64 pKa = 4.67PGLIGLNVFNEE75 pKa = 4.03TLRR78 pKa = 11.84YY79 pKa = 9.58DD80 pKa = 3.82VEE82 pKa = 4.27ILTTVYY88 pKa = 10.57EE89 pKa = 4.16KK90 pKa = 10.92DD91 pKa = 3.47ANGDD95 pKa = 3.9LQRR98 pKa = 11.84LAADD102 pKa = 4.35HH103 pKa = 6.48FPEE106 pKa = 5.37GSSANSSLTINVNVTQAKK124 pKa = 7.34TLYY127 pKa = 10.17IAVRR131 pKa = 11.84DD132 pKa = 4.04LDD134 pKa = 5.33DD135 pKa = 4.37NDD137 pKa = 4.17ASATEE142 pKa = 4.66TYY144 pKa = 10.42SISYY148 pKa = 10.3DD149 pKa = 3.34VAQAEE154 pKa = 4.53DD155 pKa = 3.75NNGSFDD161 pKa = 3.79TAVDD165 pKa = 3.94LALDD169 pKa = 4.94GACHH173 pKa = 6.52TDD175 pKa = 3.14SIGTVGDD182 pKa = 3.34VDD184 pKa = 3.74VGQFSLSNSGVYY196 pKa = 9.81EE197 pKa = 4.08ISADD201 pKa = 3.99FSAFAGGTAVDD212 pKa = 4.72LKK214 pKa = 11.33LSLFDD219 pKa = 4.88NEE221 pKa = 4.17GTLIRR226 pKa = 11.84SMGQTEE232 pKa = 4.0NSVYY236 pKa = 10.57RR237 pKa = 11.84LVEE240 pKa = 4.08SLPAGSFYY248 pKa = 11.48VLVHH252 pKa = 6.48DD253 pKa = 5.06QGKK256 pKa = 10.51DD257 pKa = 3.48DD258 pKa = 5.71FDD260 pKa = 4.0TASPFDD266 pKa = 3.88LCLSSVASAEE276 pKa = 4.0VSTDD280 pKa = 3.6DD281 pKa = 4.03NQQNATNLAGNGHH294 pKa = 5.99FEE296 pKa = 4.01ITGSIDD302 pKa = 3.31YY303 pKa = 11.03AGDD306 pKa = 3.69QDD308 pKa = 3.38WSSINSGATPPSLQVLQIEE327 pKa = 5.05FDD329 pKa = 3.64PSDD332 pKa = 4.55ANGCDD337 pKa = 2.89SWFLMEE343 pKa = 5.78VMDD346 pKa = 5.26SNDD349 pKa = 2.81VVLFSKK355 pKa = 10.44EE356 pKa = 3.9YY357 pKa = 8.97STEE360 pKa = 3.92TGPRR364 pKa = 11.84TAHH367 pKa = 6.05IRR369 pKa = 11.84VEE371 pKa = 4.19SSGEE375 pKa = 3.74HH376 pKa = 5.07FVRR379 pKa = 11.84VSGINEE385 pKa = 4.32DD386 pKa = 3.73VCSLDD391 pKa = 3.2GTVGMAYY398 pKa = 10.2SATVDD403 pKa = 3.7SVNVLDD409 pKa = 5.16DD410 pKa = 4.31AEE412 pKa = 4.82LGEE415 pKa = 4.64GNNTINTAIEE425 pKa = 4.03LDD427 pKa = 3.59EE428 pKa = 4.34TTNTEE433 pKa = 4.0TEE435 pKa = 4.01ALLSYY440 pKa = 10.6IGDD443 pKa = 3.72NDD445 pKa = 3.59WYY447 pKa = 10.84RR448 pKa = 11.84ITLPSDD454 pKa = 3.19NAQDD458 pKa = 4.0RR459 pKa = 11.84ILEE462 pKa = 4.15IFIEE466 pKa = 4.46TDD468 pKa = 2.88SEE470 pKa = 4.85TPLEE474 pKa = 4.29YY475 pKa = 10.8YY476 pKa = 9.22VTVFNADD483 pKa = 3.91EE484 pKa = 4.24IVDD487 pKa = 3.82TFTARR492 pKa = 11.84NNEE495 pKa = 3.96TSPVSFKK502 pKa = 10.94SSYY505 pKa = 10.26FIPSTAGNTNSDD517 pKa = 4.24YY518 pKa = 10.7FVKK521 pKa = 10.45IVDD524 pKa = 4.27LQSDD528 pKa = 3.71EE529 pKa = 5.39ADD531 pKa = 2.9IDD533 pKa = 3.69NNYY536 pKa = 9.45TIRR539 pKa = 11.84TNSYY543 pKa = 8.4TVQTTAPAANTGKK556 pKa = 8.46VTAAVFNSEE565 pKa = 4.19SAEE568 pKa = 4.04KK569 pKa = 10.75SLAASDD575 pKa = 3.88NTSLSLVVEE584 pKa = 4.76ASDD587 pKa = 3.41QRR589 pKa = 11.84SYY591 pKa = 11.6GVNTEE596 pKa = 3.98AFTIDD601 pKa = 3.72KK602 pKa = 8.82ATLANADD609 pKa = 3.44STITVTLPWQSGYY622 pKa = 11.25VDD624 pKa = 3.85FHH626 pKa = 7.85KK627 pKa = 11.13DD628 pKa = 3.19RR629 pKa = 11.84DD630 pKa = 3.78WFKK633 pKa = 11.67LDD635 pKa = 3.19IPTLYY640 pKa = 10.55GAAGGSAEE648 pKa = 3.82SWYY651 pKa = 11.07VEE653 pKa = 4.11VQIEE657 pKa = 4.79LYY659 pKa = 10.65SPAPGSLVEE668 pKa = 4.31YY669 pKa = 10.38AWGLYY674 pKa = 9.62RR675 pKa = 11.84DD676 pKa = 3.85EE677 pKa = 5.03SSNAVINEE685 pKa = 3.77WEE687 pKa = 4.42GEE689 pKa = 4.05DD690 pKa = 5.61GIIASNGDD698 pKa = 3.12VSALVSALDD707 pKa = 4.4LMTPAANAEE716 pKa = 4.18PMWISHH722 pKa = 5.69EE723 pKa = 4.33VANVPYY729 pKa = 9.47YY730 pKa = 10.32LTVTDD735 pKa = 5.93IINQSTNIADD745 pKa = 3.88NDD747 pKa = 3.59WGYY750 pKa = 11.22DD751 pKa = 3.08QAYY754 pKa = 9.95YY755 pKa = 10.2MRR757 pKa = 11.84AQLVFHH763 pKa = 7.16EE764 pKa = 4.93GLDD767 pKa = 3.78RR768 pKa = 11.84PADD771 pKa = 3.42
MM1 pKa = 7.53KK2 pKa = 9.98PLAQKK7 pKa = 10.04IHH9 pKa = 6.45LALLTALLLGGCGGDD24 pKa = 3.38SDD26 pKa = 5.51SPGGDD31 pKa = 2.74EE32 pKa = 5.72SGGNTNLQQNNNMTGVINPVTDD54 pKa = 3.44VDD56 pKa = 3.72WYY58 pKa = 10.6SIQVNEE64 pKa = 4.67PGLIGLNVFNEE75 pKa = 4.03TLRR78 pKa = 11.84YY79 pKa = 9.58DD80 pKa = 3.82VEE82 pKa = 4.27ILTTVYY88 pKa = 10.57EE89 pKa = 4.16KK90 pKa = 10.92DD91 pKa = 3.47ANGDD95 pKa = 3.9LQRR98 pKa = 11.84LAADD102 pKa = 4.35HH103 pKa = 6.48FPEE106 pKa = 5.37GSSANSSLTINVNVTQAKK124 pKa = 7.34TLYY127 pKa = 10.17IAVRR131 pKa = 11.84DD132 pKa = 4.04LDD134 pKa = 5.33DD135 pKa = 4.37NDD137 pKa = 4.17ASATEE142 pKa = 4.66TYY144 pKa = 10.42SISYY148 pKa = 10.3DD149 pKa = 3.34VAQAEE154 pKa = 4.53DD155 pKa = 3.75NNGSFDD161 pKa = 3.79TAVDD165 pKa = 3.94LALDD169 pKa = 4.94GACHH173 pKa = 6.52TDD175 pKa = 3.14SIGTVGDD182 pKa = 3.34VDD184 pKa = 3.74VGQFSLSNSGVYY196 pKa = 9.81EE197 pKa = 4.08ISADD201 pKa = 3.99FSAFAGGTAVDD212 pKa = 4.72LKK214 pKa = 11.33LSLFDD219 pKa = 4.88NEE221 pKa = 4.17GTLIRR226 pKa = 11.84SMGQTEE232 pKa = 4.0NSVYY236 pKa = 10.57RR237 pKa = 11.84LVEE240 pKa = 4.08SLPAGSFYY248 pKa = 11.48VLVHH252 pKa = 6.48DD253 pKa = 5.06QGKK256 pKa = 10.51DD257 pKa = 3.48DD258 pKa = 5.71FDD260 pKa = 4.0TASPFDD266 pKa = 3.88LCLSSVASAEE276 pKa = 4.0VSTDD280 pKa = 3.6DD281 pKa = 4.03NQQNATNLAGNGHH294 pKa = 5.99FEE296 pKa = 4.01ITGSIDD302 pKa = 3.31YY303 pKa = 11.03AGDD306 pKa = 3.69QDD308 pKa = 3.38WSSINSGATPPSLQVLQIEE327 pKa = 5.05FDD329 pKa = 3.64PSDD332 pKa = 4.55ANGCDD337 pKa = 2.89SWFLMEE343 pKa = 5.78VMDD346 pKa = 5.26SNDD349 pKa = 2.81VVLFSKK355 pKa = 10.44EE356 pKa = 3.9YY357 pKa = 8.97STEE360 pKa = 3.92TGPRR364 pKa = 11.84TAHH367 pKa = 6.05IRR369 pKa = 11.84VEE371 pKa = 4.19SSGEE375 pKa = 3.74HH376 pKa = 5.07FVRR379 pKa = 11.84VSGINEE385 pKa = 4.32DD386 pKa = 3.73VCSLDD391 pKa = 3.2GTVGMAYY398 pKa = 10.2SATVDD403 pKa = 3.7SVNVLDD409 pKa = 5.16DD410 pKa = 4.31AEE412 pKa = 4.82LGEE415 pKa = 4.64GNNTINTAIEE425 pKa = 4.03LDD427 pKa = 3.59EE428 pKa = 4.34TTNTEE433 pKa = 4.0TEE435 pKa = 4.01ALLSYY440 pKa = 10.6IGDD443 pKa = 3.72NDD445 pKa = 3.59WYY447 pKa = 10.84RR448 pKa = 11.84ITLPSDD454 pKa = 3.19NAQDD458 pKa = 4.0RR459 pKa = 11.84ILEE462 pKa = 4.15IFIEE466 pKa = 4.46TDD468 pKa = 2.88SEE470 pKa = 4.85TPLEE474 pKa = 4.29YY475 pKa = 10.8YY476 pKa = 9.22VTVFNADD483 pKa = 3.91EE484 pKa = 4.24IVDD487 pKa = 3.82TFTARR492 pKa = 11.84NNEE495 pKa = 3.96TSPVSFKK502 pKa = 10.94SSYY505 pKa = 10.26FIPSTAGNTNSDD517 pKa = 4.24YY518 pKa = 10.7FVKK521 pKa = 10.45IVDD524 pKa = 4.27LQSDD528 pKa = 3.71EE529 pKa = 5.39ADD531 pKa = 2.9IDD533 pKa = 3.69NNYY536 pKa = 9.45TIRR539 pKa = 11.84TNSYY543 pKa = 8.4TVQTTAPAANTGKK556 pKa = 8.46VTAAVFNSEE565 pKa = 4.19SAEE568 pKa = 4.04KK569 pKa = 10.75SLAASDD575 pKa = 3.88NTSLSLVVEE584 pKa = 4.76ASDD587 pKa = 3.41QRR589 pKa = 11.84SYY591 pKa = 11.6GVNTEE596 pKa = 3.98AFTIDD601 pKa = 3.72KK602 pKa = 8.82ATLANADD609 pKa = 3.44STITVTLPWQSGYY622 pKa = 11.25VDD624 pKa = 3.85FHH626 pKa = 7.85KK627 pKa = 11.13DD628 pKa = 3.19RR629 pKa = 11.84DD630 pKa = 3.78WFKK633 pKa = 11.67LDD635 pKa = 3.19IPTLYY640 pKa = 10.55GAAGGSAEE648 pKa = 3.82SWYY651 pKa = 11.07VEE653 pKa = 4.11VQIEE657 pKa = 4.79LYY659 pKa = 10.65SPAPGSLVEE668 pKa = 4.31YY669 pKa = 10.38AWGLYY674 pKa = 9.62RR675 pKa = 11.84DD676 pKa = 3.85EE677 pKa = 5.03SSNAVINEE685 pKa = 3.77WEE687 pKa = 4.42GEE689 pKa = 4.05DD690 pKa = 5.61GIIASNGDD698 pKa = 3.12VSALVSALDD707 pKa = 4.4LMTPAANAEE716 pKa = 4.18PMWISHH722 pKa = 5.69EE723 pKa = 4.33VANVPYY729 pKa = 9.47YY730 pKa = 10.32LTVTDD735 pKa = 5.93IINQSTNIADD745 pKa = 3.88NDD747 pKa = 3.59WGYY750 pKa = 11.22DD751 pKa = 3.08QAYY754 pKa = 9.95YY755 pKa = 10.2MRR757 pKa = 11.84AQLVFHH763 pKa = 7.16EE764 pKa = 4.93GLDD767 pKa = 3.78RR768 pKa = 11.84PADD771 pKa = 3.42
Molecular weight: 83.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y6BKT2|A0A1Y6BKT2_9ALTE Cu(I)/Ag(I) efflux system membrane protein CusA/SilA OS=Alteromonadaceae bacterium Bs31 OX=1304903 GN=SAMN02745866_00996 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.31QLAAA44 pKa = 4.14
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.31QLAAA44 pKa = 4.14
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1444979 |
18 |
5930 |
336.3 |
37.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.896 ± 0.042 | 1.089 ± 0.014 |
5.535 ± 0.044 | 6.488 ± 0.04 |
4.106 ± 0.026 | 7.129 ± 0.048 |
2.136 ± 0.019 | 5.884 ± 0.027 |
5.112 ± 0.043 | 10.235 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.238 ± 0.02 | 4.345 ± 0.036 |
4.021 ± 0.028 | 4.105 ± 0.027 |
4.768 ± 0.035 | 7.663 ± 0.069 |
5.029 ± 0.04 | 6.7 ± 0.033 |
1.354 ± 0.017 | 3.17 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
