
Apis mellifera associated microvirus 7
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
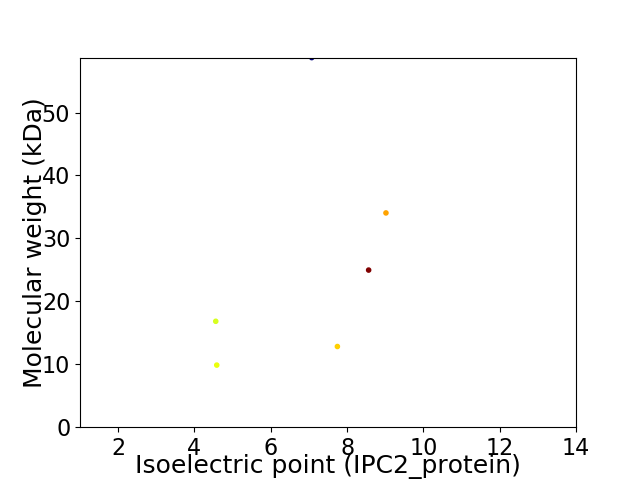
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U5A1|A0A3Q8U5A1_9VIRU Uncharacterized protein OS=Apis mellifera associated microvirus 7 OX=2494794 PE=4 SV=1
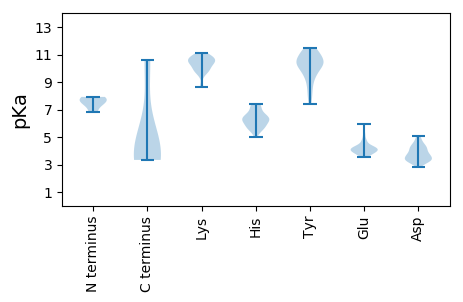
MM1 pKa = 6.79YY2 pKa = 10.31ACAVFDD8 pKa = 4.22KK9 pKa = 10.86KK10 pKa = 11.12VNAYY14 pKa = 7.72MYY16 pKa = 9.58PQFFRR21 pKa = 11.84TKK23 pKa = 10.56GEE25 pKa = 3.94ATRR28 pKa = 11.84SFMDD32 pKa = 3.34AVGAEE37 pKa = 3.71AAPFRR42 pKa = 11.84KK43 pKa = 9.42HH44 pKa = 6.84AEE46 pKa = 4.17DD47 pKa = 4.54YY48 pKa = 9.74VFCYY52 pKa = 10.27LGQYY56 pKa = 10.49DD57 pKa = 4.9DD58 pKa = 3.81NTGQFEE64 pKa = 4.33NAPGNLPEE72 pKa = 4.84ILMTAMDD79 pKa = 5.11CLTIDD84 pKa = 3.76GAVNN88 pKa = 3.44
MM1 pKa = 6.79YY2 pKa = 10.31ACAVFDD8 pKa = 4.22KK9 pKa = 10.86KK10 pKa = 11.12VNAYY14 pKa = 7.72MYY16 pKa = 9.58PQFFRR21 pKa = 11.84TKK23 pKa = 10.56GEE25 pKa = 3.94ATRR28 pKa = 11.84SFMDD32 pKa = 3.34AVGAEE37 pKa = 3.71AAPFRR42 pKa = 11.84KK43 pKa = 9.42HH44 pKa = 6.84AEE46 pKa = 4.17DD47 pKa = 4.54YY48 pKa = 9.74VFCYY52 pKa = 10.27LGQYY56 pKa = 10.49DD57 pKa = 4.9DD58 pKa = 3.81NTGQFEE64 pKa = 4.33NAPGNLPEE72 pKa = 4.84ILMTAMDD79 pKa = 5.11CLTIDD84 pKa = 3.76GAVNN88 pKa = 3.44
Molecular weight: 9.87 kDa
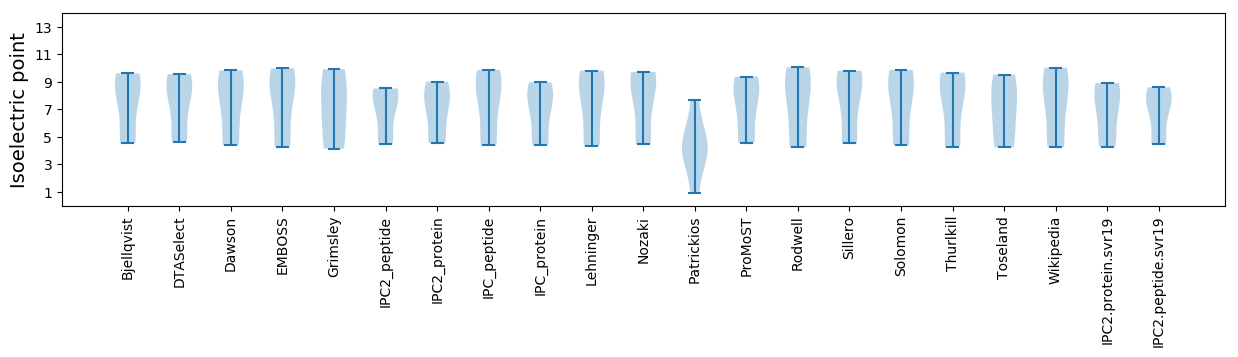
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTL2|A0A3S8UTL2_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 7 OX=2494794 PE=4 SV=1
MM1 pKa = 7.37SCYY4 pKa = 10.61GPIAAYY10 pKa = 9.87RR11 pKa = 11.84PPRR14 pKa = 11.84KK15 pKa = 9.81EE16 pKa = 4.12GGDD19 pKa = 3.28KK20 pKa = 10.73RR21 pKa = 11.84LVFDD25 pKa = 4.19PRR27 pKa = 11.84KK28 pKa = 9.83SEE30 pKa = 3.72TGVRR34 pKa = 11.84MLIPCGRR41 pKa = 11.84CTGCRR46 pKa = 11.84LEE48 pKa = 5.96RR49 pKa = 11.84SRR51 pKa = 11.84QWAMRR56 pKa = 11.84CMHH59 pKa = 7.32EE60 pKa = 4.29KK61 pKa = 10.47QMHH64 pKa = 5.76NASSFLTLTYY74 pKa = 11.13DD75 pKa = 4.63DD76 pKa = 4.31VNLPEE81 pKa = 5.1GGSLCMDD88 pKa = 4.49HH89 pKa = 6.14YY90 pKa = 11.19QKK92 pKa = 10.88FMKK95 pKa = 10.34RR96 pKa = 11.84LRR98 pKa = 11.84HH99 pKa = 5.68RR100 pKa = 11.84FGDD103 pKa = 4.4GIRR106 pKa = 11.84FCGCGEE112 pKa = 4.12YY113 pKa = 11.03GDD115 pKa = 4.1VTSRR119 pKa = 11.84PHH121 pKa = 5.02YY122 pKa = 10.47HH123 pKa = 6.95ILLLNVDD130 pKa = 4.22FGDD133 pKa = 3.34RR134 pKa = 11.84RR135 pKa = 11.84FYY137 pKa = 10.9KK138 pKa = 10.41SSEE141 pKa = 3.95GSGLPLYY148 pKa = 10.34VSKK151 pKa = 10.34EE152 pKa = 3.99LSSLWPYY159 pKa = 10.87GFSNLGDD166 pKa = 3.4VDD168 pKa = 4.85FDD170 pKa = 3.62SCAYY174 pKa = 8.0VARR177 pKa = 11.84YY178 pKa = 9.09VMKK181 pKa = 10.5KK182 pKa = 10.6VIGNDD187 pKa = 3.26TFDD190 pKa = 3.26SSLYY194 pKa = 10.14DD195 pKa = 3.34WEE197 pKa = 4.31TGIVKK202 pKa = 8.61EE203 pKa = 4.26APFMTMSRR211 pKa = 11.84RR212 pKa = 11.84PGLGSSWYY220 pKa = 10.24KK221 pKa = 10.76KK222 pKa = 10.35FGAHH226 pKa = 6.02AYY228 pKa = 8.65EE229 pKa = 4.09WDD231 pKa = 3.61SVVFKK236 pKa = 10.7GVEE239 pKa = 3.91VPPPRR244 pKa = 11.84FYY246 pKa = 11.32DD247 pKa = 3.49SLRR250 pKa = 11.84IGPAINDD257 pKa = 3.39NLKK260 pKa = 10.14KK261 pKa = 10.05KK262 pKa = 9.88RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84KK266 pKa = 9.5AALKK270 pKa = 10.52RR271 pKa = 11.84ADD273 pKa = 3.44NTLARR278 pKa = 11.84LRR280 pKa = 11.84VKK282 pKa = 10.43EE283 pKa = 4.19VVALARR289 pKa = 11.84LNLKK293 pKa = 9.87GRR295 pKa = 11.84SLL297 pKa = 3.67
MM1 pKa = 7.37SCYY4 pKa = 10.61GPIAAYY10 pKa = 9.87RR11 pKa = 11.84PPRR14 pKa = 11.84KK15 pKa = 9.81EE16 pKa = 4.12GGDD19 pKa = 3.28KK20 pKa = 10.73RR21 pKa = 11.84LVFDD25 pKa = 4.19PRR27 pKa = 11.84KK28 pKa = 9.83SEE30 pKa = 3.72TGVRR34 pKa = 11.84MLIPCGRR41 pKa = 11.84CTGCRR46 pKa = 11.84LEE48 pKa = 5.96RR49 pKa = 11.84SRR51 pKa = 11.84QWAMRR56 pKa = 11.84CMHH59 pKa = 7.32EE60 pKa = 4.29KK61 pKa = 10.47QMHH64 pKa = 5.76NASSFLTLTYY74 pKa = 11.13DD75 pKa = 4.63DD76 pKa = 4.31VNLPEE81 pKa = 5.1GGSLCMDD88 pKa = 4.49HH89 pKa = 6.14YY90 pKa = 11.19QKK92 pKa = 10.88FMKK95 pKa = 10.34RR96 pKa = 11.84LRR98 pKa = 11.84HH99 pKa = 5.68RR100 pKa = 11.84FGDD103 pKa = 4.4GIRR106 pKa = 11.84FCGCGEE112 pKa = 4.12YY113 pKa = 11.03GDD115 pKa = 4.1VTSRR119 pKa = 11.84PHH121 pKa = 5.02YY122 pKa = 10.47HH123 pKa = 6.95ILLLNVDD130 pKa = 4.22FGDD133 pKa = 3.34RR134 pKa = 11.84RR135 pKa = 11.84FYY137 pKa = 10.9KK138 pKa = 10.41SSEE141 pKa = 3.95GSGLPLYY148 pKa = 10.34VSKK151 pKa = 10.34EE152 pKa = 3.99LSSLWPYY159 pKa = 10.87GFSNLGDD166 pKa = 3.4VDD168 pKa = 4.85FDD170 pKa = 3.62SCAYY174 pKa = 8.0VARR177 pKa = 11.84YY178 pKa = 9.09VMKK181 pKa = 10.5KK182 pKa = 10.6VIGNDD187 pKa = 3.26TFDD190 pKa = 3.26SSLYY194 pKa = 10.14DD195 pKa = 3.34WEE197 pKa = 4.31TGIVKK202 pKa = 8.61EE203 pKa = 4.26APFMTMSRR211 pKa = 11.84RR212 pKa = 11.84PGLGSSWYY220 pKa = 10.24KK221 pKa = 10.76KK222 pKa = 10.35FGAHH226 pKa = 6.02AYY228 pKa = 8.65EE229 pKa = 4.09WDD231 pKa = 3.61SVVFKK236 pKa = 10.7GVEE239 pKa = 3.91VPPPRR244 pKa = 11.84FYY246 pKa = 11.32DD247 pKa = 3.49SLRR250 pKa = 11.84IGPAINDD257 pKa = 3.39NLKK260 pKa = 10.14KK261 pKa = 10.05KK262 pKa = 9.88RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84KK266 pKa = 9.5AALKK270 pKa = 10.52RR271 pKa = 11.84ADD273 pKa = 3.44NTLARR278 pKa = 11.84LRR280 pKa = 11.84VKK282 pKa = 10.43EE283 pKa = 4.19VVALARR289 pKa = 11.84LNLKK293 pKa = 9.87GRR295 pKa = 11.84SLL297 pKa = 3.67
Molecular weight: 34.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425 |
88 |
533 |
237.5 |
26.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.895 ± 1.926 | 1.684 ± 0.668 |
5.474 ± 0.442 | 4.491 ± 0.895 |
4.842 ± 0.504 | 7.579 ± 0.421 |
1.965 ± 0.265 | 3.368 ± 0.44 |
3.789 ± 0.893 | 8.281 ± 0.5 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.368 ± 0.447 | 4.842 ± 0.554 |
5.544 ± 1.011 | 3.719 ± 0.724 |
6.947 ± 0.956 | 7.93 ± 0.818 |
4.632 ± 0.697 | 7.088 ± 0.533 |
0.982 ± 0.367 | 3.579 ± 0.788 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
