
Butyricicoccus porcorum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Butyricicoccus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
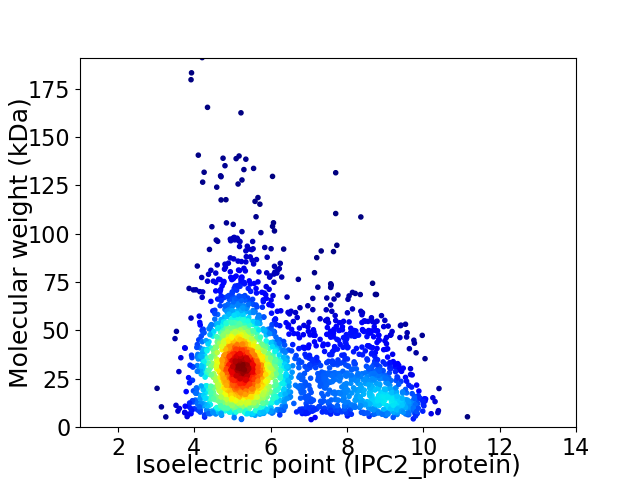
Virtual 2D-PAGE plot for 2566 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A252F4M6|A0A252F4M6_9CLOT PspC domain-containing protein OS=Butyricicoccus porcorum OX=1945634 GN=CBW42_07830 PE=4 SV=1
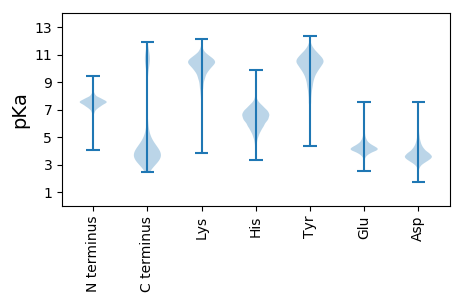
MM1 pKa = 7.31TKK3 pKa = 8.4NTKK6 pKa = 9.9RR7 pKa = 11.84LLFGVIAVAVLGGTYY22 pKa = 10.36AALLLHH28 pKa = 6.49PAEE31 pKa = 4.76EE32 pKa = 4.6EE33 pKa = 4.0ADD35 pKa = 3.83SVPSEE40 pKa = 4.36EE41 pKa = 4.52LVSMDD46 pKa = 3.56TADD49 pKa = 3.74LASVQVTLRR58 pKa = 11.84DD59 pKa = 3.64GSSFTMTTSSDD70 pKa = 3.71DD71 pKa = 3.45SGTSYY76 pKa = 10.91TMSGASEE83 pKa = 4.33DD84 pKa = 4.55DD85 pKa = 3.7YY86 pKa = 11.87SEE88 pKa = 5.27SLMQGLMDD96 pKa = 4.89AACDD100 pKa = 3.31ISARR104 pKa = 11.84PIEE107 pKa = 4.5EE108 pKa = 4.17TCSDD112 pKa = 3.77PEE114 pKa = 4.23KK115 pKa = 10.67YY116 pKa = 10.45GLSAEE121 pKa = 4.94DD122 pKa = 3.61DD123 pKa = 4.05TNTLVITDD131 pKa = 3.78TDD133 pKa = 3.83GTAVTLTIGLSGEE146 pKa = 4.08LGTYY150 pKa = 10.45CSTNGGTAVYY160 pKa = 10.62LLDD163 pKa = 4.31SDD165 pKa = 4.12TAEE168 pKa = 4.58TLTQAQSYY176 pKa = 8.34YY177 pKa = 11.05RR178 pKa = 11.84NLTVLGGYY186 pKa = 10.04YY187 pKa = 10.4SLSDD191 pKa = 3.66EE192 pKa = 4.79LKK194 pKa = 10.83SLTIDD199 pKa = 3.62HH200 pKa = 6.93MDD202 pKa = 4.54DD203 pKa = 3.42GTTVTLSARR212 pKa = 11.84DD213 pKa = 3.82TSDD216 pKa = 2.87MDD218 pKa = 4.12SDD220 pKa = 4.0VAQAYY225 pKa = 9.6SKK227 pKa = 11.26YY228 pKa = 10.9VFTAPLSCDD237 pKa = 3.42ADD239 pKa = 4.12DD240 pKa = 6.02DD241 pKa = 4.27EE242 pKa = 6.63LSTGLLSGLQSGLTAQSIVEE262 pKa = 4.74DD263 pKa = 4.01NPSDD267 pKa = 3.66LSRR270 pKa = 11.84YY271 pKa = 9.93GLDD274 pKa = 3.33DD275 pKa = 3.38PTRR278 pKa = 11.84IEE280 pKa = 4.18LAASSLDD287 pKa = 3.49AAILIGDD294 pKa = 3.9EE295 pKa = 4.21TDD297 pKa = 3.61DD298 pKa = 3.87GGIYY302 pKa = 10.29VMKK305 pKa = 10.44EE306 pKa = 3.61GGKK309 pKa = 7.65TVYY312 pKa = 10.65LCTASDD318 pKa = 3.84YY319 pKa = 11.78SFLEE323 pKa = 4.07EE324 pKa = 5.48DD325 pKa = 3.31WNDD328 pKa = 3.12WRR330 pKa = 11.84STNLLPCALSEE341 pKa = 4.08VDD343 pKa = 5.63KK344 pKa = 10.83ITVTQGDD351 pKa = 3.81EE352 pKa = 4.06THH354 pKa = 6.48TVDD357 pKa = 3.33ITHH360 pKa = 6.28VEE362 pKa = 3.81ADD364 pKa = 3.76EE365 pKa = 4.82NEE367 pKa = 4.24SDD369 pKa = 5.52DD370 pKa = 5.74ADD372 pKa = 5.21DD373 pKa = 5.16DD374 pKa = 4.25TATLDD379 pKa = 3.92GADD382 pKa = 3.62MTDD385 pKa = 3.57DD386 pKa = 3.81ALEE389 pKa = 4.06QFFLAITSVNYY400 pKa = 9.09TRR402 pKa = 11.84LIDD405 pKa = 4.48DD406 pKa = 4.71PQDD409 pKa = 3.2AAASATVTLTMNDD422 pKa = 2.83GSTRR426 pKa = 11.84SLAFTKK432 pKa = 10.52GGSRR436 pKa = 11.84EE437 pKa = 3.97YY438 pKa = 10.73LVSVDD443 pKa = 3.34GGAYY447 pKa = 10.06AYY449 pKa = 8.84GVPQDD454 pKa = 4.81DD455 pKa = 4.13LTSILDD461 pKa = 3.83ALTTGAA467 pKa = 4.93
MM1 pKa = 7.31TKK3 pKa = 8.4NTKK6 pKa = 9.9RR7 pKa = 11.84LLFGVIAVAVLGGTYY22 pKa = 10.36AALLLHH28 pKa = 6.49PAEE31 pKa = 4.76EE32 pKa = 4.6EE33 pKa = 4.0ADD35 pKa = 3.83SVPSEE40 pKa = 4.36EE41 pKa = 4.52LVSMDD46 pKa = 3.56TADD49 pKa = 3.74LASVQVTLRR58 pKa = 11.84DD59 pKa = 3.64GSSFTMTTSSDD70 pKa = 3.71DD71 pKa = 3.45SGTSYY76 pKa = 10.91TMSGASEE83 pKa = 4.33DD84 pKa = 4.55DD85 pKa = 3.7YY86 pKa = 11.87SEE88 pKa = 5.27SLMQGLMDD96 pKa = 4.89AACDD100 pKa = 3.31ISARR104 pKa = 11.84PIEE107 pKa = 4.5EE108 pKa = 4.17TCSDD112 pKa = 3.77PEE114 pKa = 4.23KK115 pKa = 10.67YY116 pKa = 10.45GLSAEE121 pKa = 4.94DD122 pKa = 3.61DD123 pKa = 4.05TNTLVITDD131 pKa = 3.78TDD133 pKa = 3.83GTAVTLTIGLSGEE146 pKa = 4.08LGTYY150 pKa = 10.45CSTNGGTAVYY160 pKa = 10.62LLDD163 pKa = 4.31SDD165 pKa = 4.12TAEE168 pKa = 4.58TLTQAQSYY176 pKa = 8.34YY177 pKa = 11.05RR178 pKa = 11.84NLTVLGGYY186 pKa = 10.04YY187 pKa = 10.4SLSDD191 pKa = 3.66EE192 pKa = 4.79LKK194 pKa = 10.83SLTIDD199 pKa = 3.62HH200 pKa = 6.93MDD202 pKa = 4.54DD203 pKa = 3.42GTTVTLSARR212 pKa = 11.84DD213 pKa = 3.82TSDD216 pKa = 2.87MDD218 pKa = 4.12SDD220 pKa = 4.0VAQAYY225 pKa = 9.6SKK227 pKa = 11.26YY228 pKa = 10.9VFTAPLSCDD237 pKa = 3.42ADD239 pKa = 4.12DD240 pKa = 6.02DD241 pKa = 4.27EE242 pKa = 6.63LSTGLLSGLQSGLTAQSIVEE262 pKa = 4.74DD263 pKa = 4.01NPSDD267 pKa = 3.66LSRR270 pKa = 11.84YY271 pKa = 9.93GLDD274 pKa = 3.33DD275 pKa = 3.38PTRR278 pKa = 11.84IEE280 pKa = 4.18LAASSLDD287 pKa = 3.49AAILIGDD294 pKa = 3.9EE295 pKa = 4.21TDD297 pKa = 3.61DD298 pKa = 3.87GGIYY302 pKa = 10.29VMKK305 pKa = 10.44EE306 pKa = 3.61GGKK309 pKa = 7.65TVYY312 pKa = 10.65LCTASDD318 pKa = 3.84YY319 pKa = 11.78SFLEE323 pKa = 4.07EE324 pKa = 5.48DD325 pKa = 3.31WNDD328 pKa = 3.12WRR330 pKa = 11.84STNLLPCALSEE341 pKa = 4.08VDD343 pKa = 5.63KK344 pKa = 10.83ITVTQGDD351 pKa = 3.81EE352 pKa = 4.06THH354 pKa = 6.48TVDD357 pKa = 3.33ITHH360 pKa = 6.28VEE362 pKa = 3.81ADD364 pKa = 3.76EE365 pKa = 4.82NEE367 pKa = 4.24SDD369 pKa = 5.52DD370 pKa = 5.74ADD372 pKa = 5.21DD373 pKa = 5.16DD374 pKa = 4.25TATLDD379 pKa = 3.92GADD382 pKa = 3.62MTDD385 pKa = 3.57DD386 pKa = 3.81ALEE389 pKa = 4.06QFFLAITSVNYY400 pKa = 9.09TRR402 pKa = 11.84LIDD405 pKa = 4.48DD406 pKa = 4.71PQDD409 pKa = 3.2AAASATVTLTMNDD422 pKa = 2.83GSTRR426 pKa = 11.84SLAFTKK432 pKa = 10.52GGSRR436 pKa = 11.84EE437 pKa = 3.97YY438 pKa = 10.73LVSVDD443 pKa = 3.34GGAYY447 pKa = 10.06AYY449 pKa = 8.84GVPQDD454 pKa = 4.81DD455 pKa = 4.13LTSILDD461 pKa = 3.83ALTTGAA467 pKa = 4.93
Molecular weight: 49.54 kDa
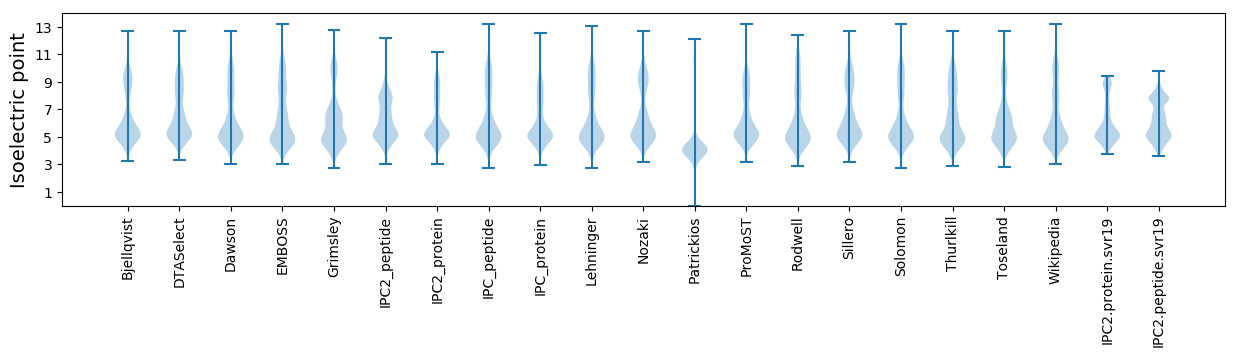
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A252F519|A0A252F519_9CLOT Bifunctional protein FolD OS=Butyricicoccus porcorum OX=1945634 GN=folD PE=3 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.8QPKK8 pKa = 9.55KK9 pKa = 7.8RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.92GFRR19 pKa = 11.84KK20 pKa = 10.08RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.83VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.15RR41 pKa = 11.84LTHH44 pKa = 6.25
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.8QPKK8 pKa = 9.55KK9 pKa = 7.8RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.92GFRR19 pKa = 11.84KK20 pKa = 10.08RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.83VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.15RR41 pKa = 11.84LTHH44 pKa = 6.25
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
777331 |
37 |
1759 |
302.9 |
33.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.252 ± 0.055 | 1.864 ± 0.025 |
5.913 ± 0.038 | 6.677 ± 0.053 |
3.794 ± 0.036 | 7.453 ± 0.046 |
2.033 ± 0.023 | 6.353 ± 0.044 |
5.051 ± 0.049 | 9.114 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.023 ± 0.022 | 3.526 ± 0.032 |
3.802 ± 0.036 | 3.611 ± 0.028 |
5.468 ± 0.061 | 5.749 ± 0.041 |
5.794 ± 0.046 | 6.988 ± 0.042 |
0.924 ± 0.016 | 3.608 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
