
Sulfitobacter sp. SK012
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Sulfitobacter; unclassified Sulfitobacter
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
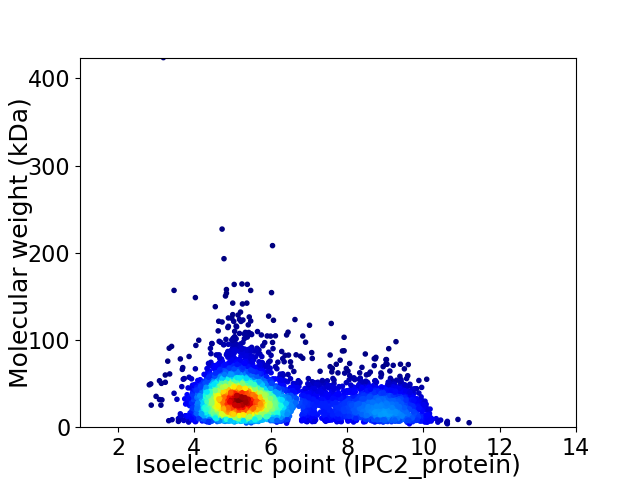
Virtual 2D-PAGE plot for 4800 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345QRS9|A0A345QRS9_9RHOB Transport permease protein OS=Sulfitobacter sp. SK012 OX=1389005 GN=C1J03_19635 PE=3 SV=1
MM1 pKa = 7.39CTMCFQLPSAPDD13 pKa = 3.38GCAFEE18 pKa = 5.02VSQPVFARR26 pKa = 11.84LRR28 pKa = 11.84EE29 pKa = 4.34GNDD32 pKa = 3.03AADD35 pKa = 4.39GVNTAYY41 pKa = 9.83TMSVGDD47 pKa = 4.43SFSGSIDD54 pKa = 3.78FNGDD58 pKa = 3.44DD59 pKa = 4.2DD60 pKa = 4.75WIAINLVSGRR70 pKa = 11.84NYY72 pKa = 10.07TISLDD77 pKa = 3.65GVTLNDD83 pKa = 3.51TYY85 pKa = 11.65LRR87 pKa = 11.84LYY89 pKa = 10.69DD90 pKa = 4.11DD91 pKa = 4.68AGSLVDD97 pKa = 5.28YY98 pKa = 11.22NDD100 pKa = 4.16DD101 pKa = 3.66ANGRR105 pKa = 11.84LDD107 pKa = 3.59SEE109 pKa = 4.52LSFRR113 pKa = 11.84LAYY116 pKa = 9.71PSGTYY121 pKa = 10.13YY122 pKa = 10.14ISAEE126 pKa = 4.12AYY128 pKa = 9.16SSFRR132 pKa = 11.84TGSYY136 pKa = 9.66EE137 pKa = 3.91LSVTGGSTPPQPPTGGAASLDD158 pKa = 3.57TLANFLTEE166 pKa = 4.56GYY168 pKa = 8.74WGGRR172 pKa = 11.84EE173 pKa = 3.64RR174 pKa = 11.84SFSTVSDD181 pKa = 4.01NIITVDD187 pKa = 3.02IGGLTSSGRR196 pKa = 11.84EE197 pKa = 3.63LARR200 pKa = 11.84WAFEE204 pKa = 3.36AWEE207 pKa = 4.22RR208 pKa = 11.84VADD211 pKa = 3.44IDD213 pKa = 4.16FRR215 pKa = 11.84EE216 pKa = 4.05VSFNADD222 pKa = 2.8IEE224 pKa = 4.42FDD226 pKa = 4.07DD227 pKa = 5.31FDD229 pKa = 6.44DD230 pKa = 4.38GAYY233 pKa = 10.59ANSQTSGGRR242 pKa = 11.84ITSSFVNVSTDD253 pKa = 2.71WLRR256 pKa = 11.84SNGTSIDD263 pKa = 3.63SYY265 pKa = 11.28SFQTYY270 pKa = 6.56VHH272 pKa = 7.14EE273 pKa = 5.04IGHH276 pKa = 6.48ALGLGHH282 pKa = 6.76QGDD285 pKa = 4.57YY286 pKa = 11.16NSNATYY292 pKa = 10.66GFDD295 pKa = 3.35EE296 pKa = 4.39NFRR299 pKa = 11.84NDD301 pKa = 2.49SWAQSVMSYY310 pKa = 10.72FDD312 pKa = 3.23QDD314 pKa = 3.41EE315 pKa = 4.37NTFTSASFARR325 pKa = 11.84LSGPMIADD333 pKa = 3.68ILAIQNLYY341 pKa = 9.91GAADD345 pKa = 4.0AGSATAGNTVYY356 pKa = 10.9GQGTTLDD363 pKa = 4.33GYY365 pKa = 10.63MGTLFDD371 pKa = 4.66ALASGRR377 pKa = 11.84TNSDD381 pKa = 3.23YY382 pKa = 10.87TGEE385 pKa = 4.78PIAFAISDD393 pKa = 3.45AGGIDD398 pKa = 4.49RR399 pKa = 11.84INLTYY404 pKa = 11.05LNGDD408 pKa = 3.29QYY410 pKa = 12.12VDD412 pKa = 3.67LRR414 pKa = 11.84QQAFSNIAGLRR425 pKa = 11.84EE426 pKa = 3.92NMSIARR432 pKa = 11.84GTTIEE437 pKa = 4.31NLNLGNGNDD446 pKa = 3.55TVYY449 pKa = 11.25GNDD452 pKa = 3.11ASNILRR458 pKa = 11.84SGGGSDD464 pKa = 5.56LLFSQNGSDD473 pKa = 3.3RR474 pKa = 11.84VFSGVGNDD482 pKa = 3.45TAYY485 pKa = 10.64GGAGNDD491 pKa = 3.67VLNGEE496 pKa = 4.55AGFDD500 pKa = 3.94QMWGGNGNDD509 pKa = 4.0YY510 pKa = 9.92LTGGAGNDD518 pKa = 3.48VLGGGGGNDD527 pKa = 3.71TLDD530 pKa = 3.65GGIGADD536 pKa = 3.27SLYY539 pKa = 11.07AGGGDD544 pKa = 3.98DD545 pKa = 4.96RR546 pKa = 11.84LLGRR550 pKa = 11.84DD551 pKa = 3.7GNDD554 pKa = 3.59NLWTSYY560 pKa = 11.77GNDD563 pKa = 3.28TVSGGNGNDD572 pKa = 3.49EE573 pKa = 4.12IGGGFGNDD581 pKa = 2.84NLYY584 pKa = 11.05AGTGNDD590 pKa = 3.28LVYY593 pKa = 10.91GGFGNDD599 pKa = 3.14VIYY602 pKa = 10.72GGTGRR607 pKa = 11.84DD608 pKa = 3.17QIFAFNQNDD617 pKa = 3.58FVDD620 pKa = 3.85AGEE623 pKa = 4.76GNDD626 pKa = 3.08IVYY629 pKa = 10.5LGRR632 pKa = 11.84GNDD635 pKa = 3.42TGRR638 pKa = 11.84GGQGDD643 pKa = 3.95DD644 pKa = 4.4AIYY647 pKa = 9.97GASGNDD653 pKa = 3.33IIAGGGGNDD662 pKa = 3.64TLTGGTGNDD671 pKa = 3.16TFIFTGGDD679 pKa = 3.19DD680 pKa = 4.75RR681 pKa = 11.84IADD684 pKa = 4.42FNASSALEE692 pKa = 4.44RR693 pKa = 11.84IDD695 pKa = 3.65FSGYY699 pKa = 10.98APISSFADD707 pKa = 3.19LRR709 pKa = 11.84SNHH712 pKa = 6.33LSSQGSNVLVNGTGGDD728 pKa = 3.4SLLILGVNLNEE739 pKa = 4.33LDD741 pKa = 3.77AGDD744 pKa = 4.09FVFF747 pKa = 5.86
MM1 pKa = 7.39CTMCFQLPSAPDD13 pKa = 3.38GCAFEE18 pKa = 5.02VSQPVFARR26 pKa = 11.84LRR28 pKa = 11.84EE29 pKa = 4.34GNDD32 pKa = 3.03AADD35 pKa = 4.39GVNTAYY41 pKa = 9.83TMSVGDD47 pKa = 4.43SFSGSIDD54 pKa = 3.78FNGDD58 pKa = 3.44DD59 pKa = 4.2DD60 pKa = 4.75WIAINLVSGRR70 pKa = 11.84NYY72 pKa = 10.07TISLDD77 pKa = 3.65GVTLNDD83 pKa = 3.51TYY85 pKa = 11.65LRR87 pKa = 11.84LYY89 pKa = 10.69DD90 pKa = 4.11DD91 pKa = 4.68AGSLVDD97 pKa = 5.28YY98 pKa = 11.22NDD100 pKa = 4.16DD101 pKa = 3.66ANGRR105 pKa = 11.84LDD107 pKa = 3.59SEE109 pKa = 4.52LSFRR113 pKa = 11.84LAYY116 pKa = 9.71PSGTYY121 pKa = 10.13YY122 pKa = 10.14ISAEE126 pKa = 4.12AYY128 pKa = 9.16SSFRR132 pKa = 11.84TGSYY136 pKa = 9.66EE137 pKa = 3.91LSVTGGSTPPQPPTGGAASLDD158 pKa = 3.57TLANFLTEE166 pKa = 4.56GYY168 pKa = 8.74WGGRR172 pKa = 11.84EE173 pKa = 3.64RR174 pKa = 11.84SFSTVSDD181 pKa = 4.01NIITVDD187 pKa = 3.02IGGLTSSGRR196 pKa = 11.84EE197 pKa = 3.63LARR200 pKa = 11.84WAFEE204 pKa = 3.36AWEE207 pKa = 4.22RR208 pKa = 11.84VADD211 pKa = 3.44IDD213 pKa = 4.16FRR215 pKa = 11.84EE216 pKa = 4.05VSFNADD222 pKa = 2.8IEE224 pKa = 4.42FDD226 pKa = 4.07DD227 pKa = 5.31FDD229 pKa = 6.44DD230 pKa = 4.38GAYY233 pKa = 10.59ANSQTSGGRR242 pKa = 11.84ITSSFVNVSTDD253 pKa = 2.71WLRR256 pKa = 11.84SNGTSIDD263 pKa = 3.63SYY265 pKa = 11.28SFQTYY270 pKa = 6.56VHH272 pKa = 7.14EE273 pKa = 5.04IGHH276 pKa = 6.48ALGLGHH282 pKa = 6.76QGDD285 pKa = 4.57YY286 pKa = 11.16NSNATYY292 pKa = 10.66GFDD295 pKa = 3.35EE296 pKa = 4.39NFRR299 pKa = 11.84NDD301 pKa = 2.49SWAQSVMSYY310 pKa = 10.72FDD312 pKa = 3.23QDD314 pKa = 3.41EE315 pKa = 4.37NTFTSASFARR325 pKa = 11.84LSGPMIADD333 pKa = 3.68ILAIQNLYY341 pKa = 9.91GAADD345 pKa = 4.0AGSATAGNTVYY356 pKa = 10.9GQGTTLDD363 pKa = 4.33GYY365 pKa = 10.63MGTLFDD371 pKa = 4.66ALASGRR377 pKa = 11.84TNSDD381 pKa = 3.23YY382 pKa = 10.87TGEE385 pKa = 4.78PIAFAISDD393 pKa = 3.45AGGIDD398 pKa = 4.49RR399 pKa = 11.84INLTYY404 pKa = 11.05LNGDD408 pKa = 3.29QYY410 pKa = 12.12VDD412 pKa = 3.67LRR414 pKa = 11.84QQAFSNIAGLRR425 pKa = 11.84EE426 pKa = 3.92NMSIARR432 pKa = 11.84GTTIEE437 pKa = 4.31NLNLGNGNDD446 pKa = 3.55TVYY449 pKa = 11.25GNDD452 pKa = 3.11ASNILRR458 pKa = 11.84SGGGSDD464 pKa = 5.56LLFSQNGSDD473 pKa = 3.3RR474 pKa = 11.84VFSGVGNDD482 pKa = 3.45TAYY485 pKa = 10.64GGAGNDD491 pKa = 3.67VLNGEE496 pKa = 4.55AGFDD500 pKa = 3.94QMWGGNGNDD509 pKa = 4.0YY510 pKa = 9.92LTGGAGNDD518 pKa = 3.48VLGGGGGNDD527 pKa = 3.71TLDD530 pKa = 3.65GGIGADD536 pKa = 3.27SLYY539 pKa = 11.07AGGGDD544 pKa = 3.98DD545 pKa = 4.96RR546 pKa = 11.84LLGRR550 pKa = 11.84DD551 pKa = 3.7GNDD554 pKa = 3.59NLWTSYY560 pKa = 11.77GNDD563 pKa = 3.28TVSGGNGNDD572 pKa = 3.49EE573 pKa = 4.12IGGGFGNDD581 pKa = 2.84NLYY584 pKa = 11.05AGTGNDD590 pKa = 3.28LVYY593 pKa = 10.91GGFGNDD599 pKa = 3.14VIYY602 pKa = 10.72GGTGRR607 pKa = 11.84DD608 pKa = 3.17QIFAFNQNDD617 pKa = 3.58FVDD620 pKa = 3.85AGEE623 pKa = 4.76GNDD626 pKa = 3.08IVYY629 pKa = 10.5LGRR632 pKa = 11.84GNDD635 pKa = 3.42TGRR638 pKa = 11.84GGQGDD643 pKa = 3.95DD644 pKa = 4.4AIYY647 pKa = 9.97GASGNDD653 pKa = 3.33IIAGGGGNDD662 pKa = 3.64TLTGGTGNDD671 pKa = 3.16TFIFTGGDD679 pKa = 3.19DD680 pKa = 4.75RR681 pKa = 11.84IADD684 pKa = 4.42FNASSALEE692 pKa = 4.44RR693 pKa = 11.84IDD695 pKa = 3.65FSGYY699 pKa = 10.98APISSFADD707 pKa = 3.19LRR709 pKa = 11.84SNHH712 pKa = 6.33LSSQGSNVLVNGTGGDD728 pKa = 3.4SLLILGVNLNEE739 pKa = 4.33LDD741 pKa = 3.77AGDD744 pKa = 4.09FVFF747 pKa = 5.86
Molecular weight: 78.29 kDa
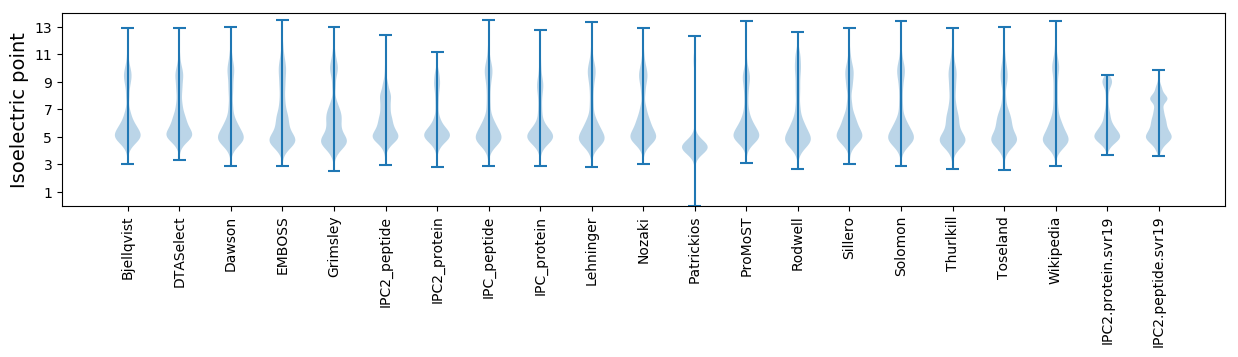
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345QIY8|A0A345QIY8_9RHOB Multidrug ABC transporter ATP-binding protein OS=Sulfitobacter sp. SK012 OX=1389005 GN=C1J03_04025 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84SKK41 pKa = 11.13LSAA44 pKa = 3.7
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84SKK41 pKa = 11.13LSAA44 pKa = 3.7
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1475871 |
34 |
4097 |
307.5 |
33.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.556 ± 0.049 | 0.916 ± 0.013 |
6.135 ± 0.036 | 5.572 ± 0.036 |
3.934 ± 0.025 | 8.518 ± 0.037 |
2.086 ± 0.018 | 5.555 ± 0.025 |
3.521 ± 0.031 | 9.866 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.905 ± 0.017 | 2.988 ± 0.021 |
4.857 ± 0.026 | 3.384 ± 0.019 |
5.992 ± 0.033 | 5.613 ± 0.024 |
5.664 ± 0.028 | 7.219 ± 0.03 |
1.416 ± 0.016 | 2.305 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
