
Microbacterium sp. ABRD_28
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
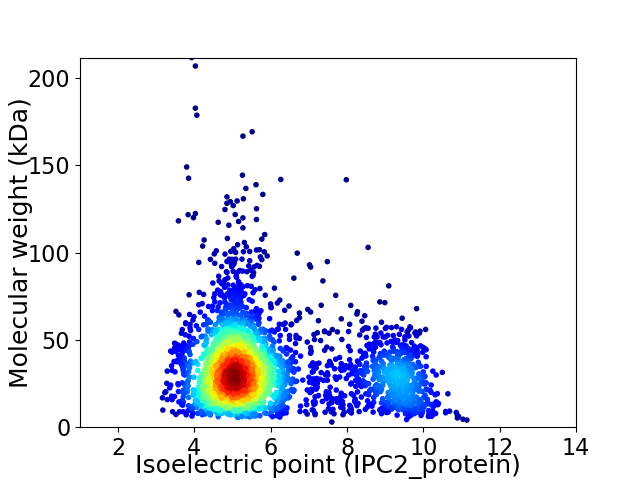
Virtual 2D-PAGE plot for 3036 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G6ZJK0|A0A3G6ZJK0_9MICO Rho termination factor OS=Microbacterium sp. ABRD_28 OX=2268461 GN=DT073_03550 PE=4 SV=1
MM1 pKa = 7.59SNTPEE6 pKa = 5.02PDD8 pKa = 4.18DD9 pKa = 4.5SPQGPTYY16 pKa = 10.06EE17 pKa = 4.29GRR19 pKa = 11.84VLPRR23 pKa = 11.84PEE25 pKa = 5.21DD26 pKa = 3.58EE27 pKa = 5.52VVDD30 pKa = 4.0QGAGFDD36 pKa = 3.71VATMVSRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84VLTLVGLGVGAATLAACAATTDD67 pKa = 4.1SPTSATSTATTDD79 pKa = 3.28STATEE84 pKa = 4.3ATDD87 pKa = 3.42STTTTTAAGEE97 pKa = 4.24IPEE100 pKa = 4.53EE101 pKa = 4.35TNGPYY106 pKa = 9.97PADD109 pKa = 3.63GSNGVNVLEE118 pKa = 4.14EE119 pKa = 4.19SGIVRR124 pKa = 11.84SDD126 pKa = 2.76IRR128 pKa = 11.84SSLDD132 pKa = 2.92GGTTAEE138 pKa = 4.3GVPLTFTFSVTDD150 pKa = 3.44IAGGDD155 pKa = 3.69VPFEE159 pKa = 4.1GAAVYY164 pKa = 9.72VWHH167 pKa = 7.64CDD169 pKa = 3.38AQGLYY174 pKa = 10.99SMYY177 pKa = 10.74SEE179 pKa = 4.2GVEE182 pKa = 4.53DD183 pKa = 3.69EE184 pKa = 4.36TFLRR188 pKa = 11.84GIQVVDD194 pKa = 3.58ARR196 pKa = 11.84GEE198 pKa = 4.14ATFQTIVPGCYY209 pKa = 9.22AGRR212 pKa = 11.84WTHH215 pKa = 5.54IHH217 pKa = 6.19FEE219 pKa = 4.36VYY221 pKa = 10.25PDD223 pKa = 3.18IASATDD229 pKa = 3.4VANVIATSQVAFPEE243 pKa = 4.11EE244 pKa = 4.11MLNAVYY250 pKa = 10.42QLDD253 pKa = 4.35AYY255 pKa = 10.82SGSAQNLAQIGGLANDD271 pKa = 4.08NVFSDD276 pKa = 3.84GTEE279 pKa = 3.84LQMGTFSGDD288 pKa = 2.98TSSGYY293 pKa = 10.96VGTLAVGVDD302 pKa = 3.67TDD304 pKa = 3.84TAPGVSAGAGGAGGGQPPSGGGPGNN329 pKa = 3.49
MM1 pKa = 7.59SNTPEE6 pKa = 5.02PDD8 pKa = 4.18DD9 pKa = 4.5SPQGPTYY16 pKa = 10.06EE17 pKa = 4.29GRR19 pKa = 11.84VLPRR23 pKa = 11.84PEE25 pKa = 5.21DD26 pKa = 3.58EE27 pKa = 5.52VVDD30 pKa = 4.0QGAGFDD36 pKa = 3.71VATMVSRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84VLTLVGLGVGAATLAACAATTDD67 pKa = 4.1SPTSATSTATTDD79 pKa = 3.28STATEE84 pKa = 4.3ATDD87 pKa = 3.42STTTTTAAGEE97 pKa = 4.24IPEE100 pKa = 4.53EE101 pKa = 4.35TNGPYY106 pKa = 9.97PADD109 pKa = 3.63GSNGVNVLEE118 pKa = 4.14EE119 pKa = 4.19SGIVRR124 pKa = 11.84SDD126 pKa = 2.76IRR128 pKa = 11.84SSLDD132 pKa = 2.92GGTTAEE138 pKa = 4.3GVPLTFTFSVTDD150 pKa = 3.44IAGGDD155 pKa = 3.69VPFEE159 pKa = 4.1GAAVYY164 pKa = 9.72VWHH167 pKa = 7.64CDD169 pKa = 3.38AQGLYY174 pKa = 10.99SMYY177 pKa = 10.74SEE179 pKa = 4.2GVEE182 pKa = 4.53DD183 pKa = 3.69EE184 pKa = 4.36TFLRR188 pKa = 11.84GIQVVDD194 pKa = 3.58ARR196 pKa = 11.84GEE198 pKa = 4.14ATFQTIVPGCYY209 pKa = 9.22AGRR212 pKa = 11.84WTHH215 pKa = 5.54IHH217 pKa = 6.19FEE219 pKa = 4.36VYY221 pKa = 10.25PDD223 pKa = 3.18IASATDD229 pKa = 3.4VANVIATSQVAFPEE243 pKa = 4.11EE244 pKa = 4.11MLNAVYY250 pKa = 10.42QLDD253 pKa = 4.35AYY255 pKa = 10.82SGSAQNLAQIGGLANDD271 pKa = 4.08NVFSDD276 pKa = 3.84GTEE279 pKa = 3.84LQMGTFSGDD288 pKa = 2.98TSSGYY293 pKa = 10.96VGTLAVGVDD302 pKa = 3.67TDD304 pKa = 3.84TAPGVSAGAGGAGGGQPPSGGGPGNN329 pKa = 3.49
Molecular weight: 33.41 kDa
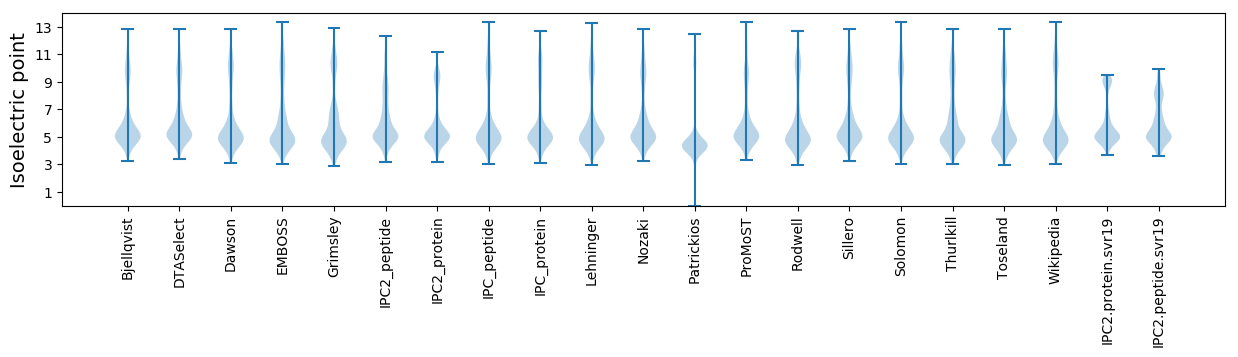
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G6ZQR2|A0A3G6ZQR2_9MICO Neutral metalloproteinase OS=Microbacterium sp. ABRD_28 OX=2268461 GN=DT073_07260 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
994539 |
29 |
2004 |
327.6 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.778 ± 0.067 | 0.481 ± 0.01 |
6.464 ± 0.038 | 5.685 ± 0.041 |
3.156 ± 0.028 | 9.042 ± 0.042 |
1.933 ± 0.023 | 4.5 ± 0.031 |
1.577 ± 0.029 | 10.08 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.708 ± 0.016 | 1.733 ± 0.023 |
5.61 ± 0.031 | 2.592 ± 0.022 |
7.821 ± 0.051 | 5.324 ± 0.028 |
5.96 ± 0.033 | 9.126 ± 0.04 |
1.538 ± 0.023 | 1.891 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
