
Cellulosimicrobium cellulans J1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Cellulosimicrobium; Cellulosimicrobium cellulans
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
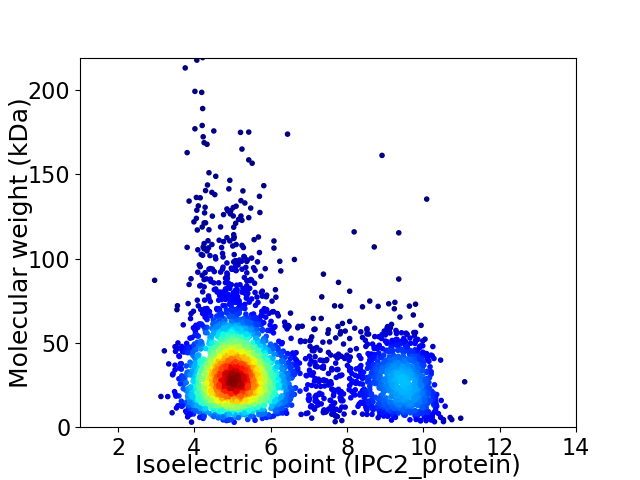
Virtual 2D-PAGE plot for 3743 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
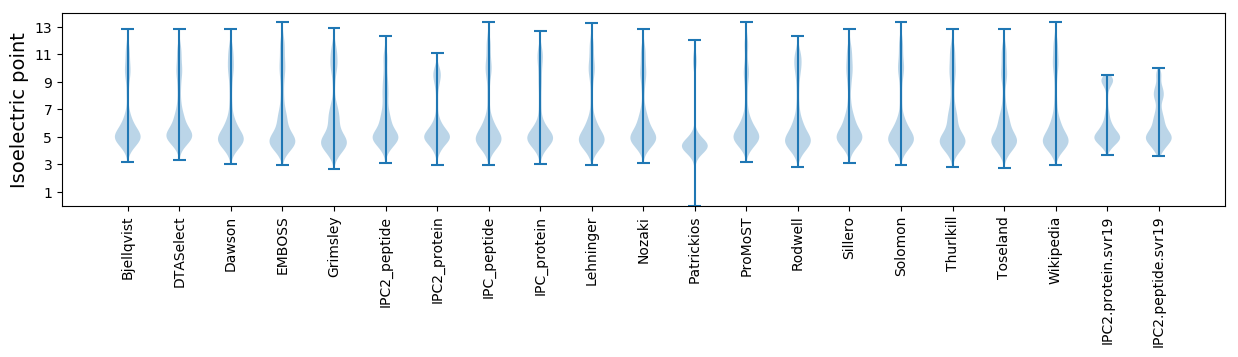
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7DMU4|A0A1X7DMU4_CELCE Beta-N-acetylhexosaminidase OS=Cellulosimicrobium cellulans J1 OX=903523 GN=SAMN02744115_01861 PE=3 SV=1
MM1 pKa = 7.51ALVTAGSGLAAAAAPANAATVYY23 pKa = 9.46EE24 pKa = 5.04LEE26 pKa = 4.65GAWSAGTPTTVATGDD41 pKa = 3.59AVSASWWFNLNDD53 pKa = 4.1DD54 pKa = 4.26AAAPGNAPVDD64 pKa = 3.86DD65 pKa = 3.84VTITVTGQGAVFDD78 pKa = 5.2SIPPVCLTDD87 pKa = 3.93GVDD90 pKa = 3.61PASSISDD97 pKa = 3.52DD98 pKa = 4.13GATLVCNVGTQDD110 pKa = 3.31EE111 pKa = 4.83GTAFALTTPMRR122 pKa = 11.84VTAQTGEE129 pKa = 4.23PVSAGATVADD139 pKa = 4.23QEE141 pKa = 4.48ATVPDD146 pKa = 4.42LVVQNAFLQDD156 pKa = 3.58IRR158 pKa = 11.84WVQATTQSGNTATYY172 pKa = 10.75RR173 pKa = 11.84DD174 pKa = 3.99MVFNWTLFHH183 pKa = 6.43GTHH186 pKa = 6.11SPAGPSEE193 pKa = 3.99VTYY196 pKa = 11.34ALTLSNTLGATMQLGPDD213 pKa = 3.79GCRR216 pKa = 11.84AFTSGSASGHH226 pKa = 5.77PWSGGSNDD234 pKa = 4.0PSQTAPFVGTCTLTRR249 pKa = 11.84NSSTSYY255 pKa = 11.25SLTLSGIDD263 pKa = 3.61YY264 pKa = 10.89SHH266 pKa = 6.39TQVPTKK272 pKa = 10.78DD273 pKa = 3.27SAGANLPTDD282 pKa = 3.9RR283 pKa = 11.84VAVASGQVQIRR294 pKa = 11.84VMTASNSGGITLTSNAPTYY313 pKa = 9.7TAVDD317 pKa = 3.87GQTYY321 pKa = 10.06TDD323 pKa = 3.74DD324 pKa = 3.77ASNNSVNRR332 pKa = 11.84TWTAGYY338 pKa = 8.59WSHH341 pKa = 6.38AWTPAYY347 pKa = 9.11TGQSGSSWSDD357 pKa = 3.03TYY359 pKa = 10.95RR360 pKa = 11.84VAAGTEE366 pKa = 4.13VSSFASVNYY375 pKa = 9.91PNVSTPNATQGGLCVVLDD393 pKa = 3.89TQNVEE398 pKa = 4.09FTGATTRR405 pKa = 11.84FDD407 pKa = 3.85NNRR410 pKa = 11.84TGPEE414 pKa = 3.63AGLPLAYY421 pKa = 9.35YY422 pKa = 9.26TGTSALLNPDD432 pKa = 3.39SGSYY436 pKa = 9.59QPNDD440 pKa = 3.38FTCNGTTGWTTTRR453 pKa = 11.84PTDD456 pKa = 3.42LSTIKK461 pKa = 10.5AVRR464 pKa = 11.84TVFTPTNAIKK474 pKa = 10.33EE475 pKa = 4.3RR476 pKa = 11.84LVALQVTQRR485 pKa = 11.84VKK487 pKa = 9.62EE488 pKa = 4.25TTPVGRR494 pKa = 11.84DD495 pKa = 2.72VWTWGAYY502 pKa = 8.71INAVNTSNVWVHH514 pKa = 6.27PNRR517 pKa = 11.84SSDD520 pKa = 3.67TADD523 pKa = 3.12MPLNGTATPGSRR535 pKa = 11.84YY536 pKa = 10.06AYY538 pKa = 9.86AAGGRR543 pKa = 11.84DD544 pKa = 3.69VLRR547 pKa = 11.84VIGVTPDD554 pKa = 2.91VRR556 pKa = 11.84KK557 pKa = 9.93SVLPITVDD565 pKa = 3.37PSGEE569 pKa = 3.76ATYY572 pKa = 10.49TLTYY576 pKa = 9.71SANGTGAVAPTVDD589 pKa = 5.23DD590 pKa = 3.95YY591 pKa = 12.31VLVDD595 pKa = 4.0TLPAGATYY603 pKa = 10.82VGGSASPEE611 pKa = 3.86PAVTTNGSGQQVLTWTLDD629 pKa = 3.51GVATNAEE636 pKa = 4.18HH637 pKa = 6.16SLSYY641 pKa = 10.46VVRR644 pKa = 11.84FPDD647 pKa = 4.15DD648 pKa = 3.49LQGGDD653 pKa = 3.21RR654 pKa = 11.84LVNTVSAAVEE664 pKa = 4.5GVTSDD669 pKa = 3.81DD670 pKa = 3.29ATASVVVSDD679 pKa = 4.58SGLTKK684 pKa = 10.02IIKK687 pKa = 8.82TADD690 pKa = 3.09QAFIPNLAGDD700 pKa = 4.34GVGTGSWTVLVRR712 pKa = 11.84SQDD715 pKa = 3.85PIAQSFVDD723 pKa = 4.98VIDD726 pKa = 3.49ILPYY730 pKa = 10.33VGDD733 pKa = 3.75GRR735 pKa = 11.84GTAFSGSYY743 pKa = 10.68LLDD746 pKa = 3.98GPVTSSIGGTVYY758 pKa = 9.49YY759 pKa = 7.49TTADD763 pKa = 3.77PTTLTDD769 pKa = 5.21DD770 pKa = 4.48PADD773 pKa = 4.2PSNGSAGSVAGNTVGWTTEE792 pKa = 3.93YY793 pKa = 10.77TPDD796 pKa = 3.1ATAVRR801 pKa = 11.84VITGTLAPRR810 pKa = 11.84GEE812 pKa = 4.36LTISVPVVTDD822 pKa = 3.21GMVGGDD828 pKa = 3.36TLVNRR833 pKa = 11.84SQGRR837 pKa = 11.84AEE839 pKa = 3.9NTRR842 pKa = 11.84LVMRR846 pKa = 11.84TSAPTSIANYY856 pKa = 10.02YY857 pKa = 9.47SASLKK862 pKa = 10.66KK863 pKa = 10.44YY864 pKa = 9.53VQDD867 pKa = 5.32ADD869 pKa = 4.43GEE871 pKa = 4.37WRR873 pKa = 11.84DD874 pKa = 3.72ANDD877 pKa = 3.96PLDD880 pKa = 3.91YY881 pKa = 10.67PQFRR885 pKa = 11.84VGDD888 pKa = 3.75TVRR891 pKa = 11.84YY892 pKa = 9.3RR893 pKa = 11.84IVVEE897 pKa = 3.83NTGQGTLTNVVVSDD911 pKa = 4.21DD912 pKa = 4.09LQPEE916 pKa = 4.52LGSFTIEE923 pKa = 3.76SLAPGEE929 pKa = 4.43VEE931 pKa = 3.81SHH933 pKa = 5.59EE934 pKa = 4.23FEE936 pKa = 4.35IVLSEE941 pKa = 4.11PVDD944 pKa = 3.67DD945 pKa = 4.79AVVNTACADD954 pKa = 3.47ADD956 pKa = 3.81VPEE959 pKa = 5.03DD960 pKa = 3.61SGVEE964 pKa = 3.82PTINCDD970 pKa = 3.08PAGFTVVGTPTHH982 pKa = 5.34TKK984 pKa = 9.57EE985 pKa = 4.21IVSATPVGDD994 pKa = 3.73GRR996 pKa = 11.84WEE998 pKa = 3.86VVYY1001 pKa = 10.24EE1002 pKa = 4.08VVVSNTQTPSTTYY1015 pKa = 10.84SLTDD1019 pKa = 3.4EE1020 pKa = 5.01LHH1022 pKa = 6.03FTDD1025 pKa = 3.54QATIVSATVTQAPDD1039 pKa = 3.58GVVLADD1045 pKa = 4.12PAWDD1049 pKa = 3.67GQEE1052 pKa = 4.05NLTIASDD1059 pKa = 3.94VPLAGNDD1066 pKa = 3.44DD1067 pKa = 4.44DD1068 pKa = 6.15GYY1070 pKa = 9.77EE1071 pKa = 3.86PHH1073 pKa = 6.83VYY1075 pKa = 9.82RR1076 pKa = 11.84VTVLADD1082 pKa = 3.21VPLQLDD1088 pKa = 4.16GAGSGADD1095 pKa = 4.74DD1096 pKa = 3.83PTACPAEE1103 pKa = 4.67GSDD1106 pKa = 4.92ADD1108 pKa = 3.46QGFNNTSGLTGPDD1121 pKa = 3.52GEE1123 pKa = 4.59TEE1125 pKa = 4.02EE1126 pKa = 5.26DD1127 pKa = 3.73QACAPIPSIDD1137 pKa = 3.44VEE1139 pKa = 4.34KK1140 pKa = 10.55RR1141 pKa = 11.84VAAGPTPNGDD1151 pKa = 3.68GTWTVTYY1158 pKa = 10.57DD1159 pKa = 3.59VVATNSGTGDD1169 pKa = 3.25GVYY1172 pKa = 10.42EE1173 pKa = 4.15VTDD1176 pKa = 3.49RR1177 pKa = 11.84MTADD1181 pKa = 3.33GDD1183 pKa = 4.17LEE1185 pKa = 4.6VVSGAVTSAPDD1196 pKa = 3.62GVTPSATWTGLGAEE1210 pKa = 4.51GAPEE1214 pKa = 3.87NVIASAVTLPAGTSHH1229 pKa = 7.15AYY1231 pKa = 8.88EE1232 pKa = 4.18VQVVLTIAPGTEE1244 pKa = 3.95GAPVVTPCSAEE1255 pKa = 3.86PGGSSGGLSNAAGVEE1270 pKa = 4.35HH1271 pKa = 7.47NDD1273 pKa = 3.45LTDD1276 pKa = 3.28TAEE1279 pKa = 4.23ACVTLAAITVEE1290 pKa = 4.4KK1291 pKa = 10.24SISAGPTPNGDD1302 pKa = 3.71GTWTVVYY1309 pKa = 10.37DD1310 pKa = 4.06VVAEE1314 pKa = 4.16NVGAAAGEE1322 pKa = 3.96YY1323 pKa = 10.18DD1324 pKa = 3.48VYY1326 pKa = 11.69DD1327 pKa = 3.63RR1328 pKa = 11.84LRR1330 pKa = 11.84YY1331 pKa = 9.77GDD1333 pKa = 4.22GIEE1336 pKa = 4.2IVDD1339 pKa = 3.89AAVTSGPDD1347 pKa = 3.39GVTTNAAWTGLGEE1360 pKa = 4.78PDD1362 pKa = 3.09TAAEE1366 pKa = 4.15NLVAADD1372 pKa = 3.7VEE1374 pKa = 4.48LAAGGLHH1381 pKa = 5.92TFQVEE1386 pKa = 4.13VTVRR1390 pKa = 11.84MDD1392 pKa = 3.22EE1393 pKa = 4.03ATIDD1397 pKa = 3.94PVALRR1402 pKa = 11.84CPPPGSGASGGLANSTSLDD1421 pKa = 3.39HH1422 pKa = 6.64NGIVAEE1428 pKa = 4.69DD1429 pKa = 4.23EE1430 pKa = 4.67VCASLPLIGVEE1441 pKa = 4.39KK1442 pKa = 10.27SISAGPVPQGDD1453 pKa = 4.2GTWAITYY1460 pKa = 10.48DD1461 pKa = 4.51LVATNAGEE1469 pKa = 3.96AAGDD1473 pKa = 3.59YY1474 pKa = 11.2DD1475 pKa = 5.31LVDD1478 pKa = 3.49RR1479 pKa = 11.84LQYY1482 pKa = 10.91GEE1484 pKa = 4.3GVVVEE1489 pKa = 4.48SAAVVSAPDD1498 pKa = 3.83GVATEE1503 pKa = 4.8AGWTGQGAAGSTEE1516 pKa = 3.95NVVAAGVTLGAGAAHH1531 pKa = 7.38TYY1533 pKa = 8.7QVQVVVSLDD1542 pKa = 3.48RR1543 pKa = 11.84AVVTPSTLACPEE1555 pKa = 4.54PGSGEE1560 pKa = 4.16GGGLANATEE1569 pKa = 4.29LTHH1572 pKa = 7.49NGEE1575 pKa = 4.31TQGDD1579 pKa = 4.21DD1580 pKa = 3.41VCAEE1584 pKa = 4.43LPLIDD1589 pKa = 3.88IAKK1592 pKa = 8.81SLSGAVTPVEE1602 pKa = 4.32GEE1604 pKa = 3.9PGVYY1608 pKa = 10.07DD1609 pKa = 3.68AVYY1612 pKa = 10.34EE1613 pKa = 4.17ITVTNAGPGDD1623 pKa = 3.97GLYY1626 pKa = 11.08DD1627 pKa = 3.93LTDD1630 pKa = 3.39EE1631 pKa = 4.6LAPGEE1636 pKa = 4.23GVEE1639 pKa = 4.15VLGVQDD1645 pKa = 4.0VVTDD1649 pKa = 3.75APEE1652 pKa = 3.94PVGLNPGFDD1661 pKa = 3.71GTSDD1665 pKa = 3.6TLVVAGQPIAGATTAPVVHH1684 pKa = 7.1TYY1686 pKa = 9.23TVTVRR1691 pKa = 11.84YY1692 pKa = 9.04AADD1695 pKa = 3.72LSDD1698 pKa = 4.69VEE1700 pKa = 5.49IPDD1703 pKa = 3.63TDD1705 pKa = 3.28ACTTGDD1711 pKa = 3.45GSTVAGGLANTATVGWNGLDD1731 pKa = 3.76GTDD1734 pKa = 4.14DD1735 pKa = 3.54ACVRR1739 pKa = 11.84PGKK1742 pKa = 8.38PTLDD1746 pKa = 3.44KK1747 pKa = 11.02ALVSAAPAGDD1757 pKa = 3.84GTWEE1761 pKa = 4.04VVYY1764 pKa = 10.66DD1765 pKa = 3.7LTVGNVGGEE1774 pKa = 3.86ATTYY1778 pKa = 11.33DD1779 pKa = 4.64LDD1781 pKa = 5.91DD1782 pKa = 4.55EE1783 pKa = 5.49LLFAPVVTVDD1793 pKa = 3.56SVAVTGPEE1801 pKa = 3.91GVTVNAGFDD1810 pKa = 3.34GATDD1814 pKa = 3.43QRR1816 pKa = 11.84IATGVSIAGMDD1827 pKa = 3.73DD1828 pKa = 3.11EE1829 pKa = 6.21GYY1831 pKa = 10.42APHH1834 pKa = 7.05VYY1836 pKa = 9.69RR1837 pKa = 11.84VTVVAHH1843 pKa = 5.84VPLSFAADD1851 pKa = 3.73DD1852 pKa = 3.8VDD1854 pKa = 4.83GDD1856 pKa = 4.33GTGSPACTVPAGSNTLQQGLNNAATLTDD1884 pKa = 3.88EE1885 pKa = 4.55TGGTQTDD1892 pKa = 3.89TDD1894 pKa = 4.43CAPLPSIDD1902 pKa = 3.66VTKK1905 pKa = 10.33STDD1908 pKa = 3.27GDD1910 pKa = 4.0PVQGDD1915 pKa = 3.61DD1916 pKa = 4.03GSWTVRR1922 pKa = 11.84YY1923 pKa = 9.43AVTVTNDD1930 pKa = 2.98GGSAGAYY1937 pKa = 10.1DD1938 pKa = 3.59LTDD1941 pKa = 3.5RR1942 pKa = 11.84LRR1944 pKa = 11.84YY1945 pKa = 8.97GAGITVRR1952 pKa = 11.84SATVTATPEE1961 pKa = 4.09GVTASATWTGQGAEE1975 pKa = 3.8GDD1977 pKa = 3.81AANVVAADD1985 pKa = 3.42VDD1987 pKa = 4.35LAPGATHH1994 pKa = 6.66VYY1996 pKa = 9.41RR1997 pKa = 11.84VEE1999 pKa = 3.96VVAAVDD2005 pKa = 3.76PGRR2008 pKa = 11.84ADD2010 pKa = 3.07ATTFTCPAPGSGEE2023 pKa = 3.81AGGFANTAGVGHH2035 pKa = 7.01NDD2037 pKa = 3.09LTDD2040 pKa = 3.84AADD2043 pKa = 3.48ACAAPPEE2050 pKa = 4.38PANPGTPGKK2059 pKa = 9.67PGGSLATTGATLAWVAGGAALLLLLGMLALLAARR2093 pKa = 11.84RR2094 pKa = 11.84RR2095 pKa = 11.84RR2096 pKa = 11.84LTSS2099 pKa = 2.99
MM1 pKa = 7.51ALVTAGSGLAAAAAPANAATVYY23 pKa = 9.46EE24 pKa = 5.04LEE26 pKa = 4.65GAWSAGTPTTVATGDD41 pKa = 3.59AVSASWWFNLNDD53 pKa = 4.1DD54 pKa = 4.26AAAPGNAPVDD64 pKa = 3.86DD65 pKa = 3.84VTITVTGQGAVFDD78 pKa = 5.2SIPPVCLTDD87 pKa = 3.93GVDD90 pKa = 3.61PASSISDD97 pKa = 3.52DD98 pKa = 4.13GATLVCNVGTQDD110 pKa = 3.31EE111 pKa = 4.83GTAFALTTPMRR122 pKa = 11.84VTAQTGEE129 pKa = 4.23PVSAGATVADD139 pKa = 4.23QEE141 pKa = 4.48ATVPDD146 pKa = 4.42LVVQNAFLQDD156 pKa = 3.58IRR158 pKa = 11.84WVQATTQSGNTATYY172 pKa = 10.75RR173 pKa = 11.84DD174 pKa = 3.99MVFNWTLFHH183 pKa = 6.43GTHH186 pKa = 6.11SPAGPSEE193 pKa = 3.99VTYY196 pKa = 11.34ALTLSNTLGATMQLGPDD213 pKa = 3.79GCRR216 pKa = 11.84AFTSGSASGHH226 pKa = 5.77PWSGGSNDD234 pKa = 4.0PSQTAPFVGTCTLTRR249 pKa = 11.84NSSTSYY255 pKa = 11.25SLTLSGIDD263 pKa = 3.61YY264 pKa = 10.89SHH266 pKa = 6.39TQVPTKK272 pKa = 10.78DD273 pKa = 3.27SAGANLPTDD282 pKa = 3.9RR283 pKa = 11.84VAVASGQVQIRR294 pKa = 11.84VMTASNSGGITLTSNAPTYY313 pKa = 9.7TAVDD317 pKa = 3.87GQTYY321 pKa = 10.06TDD323 pKa = 3.74DD324 pKa = 3.77ASNNSVNRR332 pKa = 11.84TWTAGYY338 pKa = 8.59WSHH341 pKa = 6.38AWTPAYY347 pKa = 9.11TGQSGSSWSDD357 pKa = 3.03TYY359 pKa = 10.95RR360 pKa = 11.84VAAGTEE366 pKa = 4.13VSSFASVNYY375 pKa = 9.91PNVSTPNATQGGLCVVLDD393 pKa = 3.89TQNVEE398 pKa = 4.09FTGATTRR405 pKa = 11.84FDD407 pKa = 3.85NNRR410 pKa = 11.84TGPEE414 pKa = 3.63AGLPLAYY421 pKa = 9.35YY422 pKa = 9.26TGTSALLNPDD432 pKa = 3.39SGSYY436 pKa = 9.59QPNDD440 pKa = 3.38FTCNGTTGWTTTRR453 pKa = 11.84PTDD456 pKa = 3.42LSTIKK461 pKa = 10.5AVRR464 pKa = 11.84TVFTPTNAIKK474 pKa = 10.33EE475 pKa = 4.3RR476 pKa = 11.84LVALQVTQRR485 pKa = 11.84VKK487 pKa = 9.62EE488 pKa = 4.25TTPVGRR494 pKa = 11.84DD495 pKa = 2.72VWTWGAYY502 pKa = 8.71INAVNTSNVWVHH514 pKa = 6.27PNRR517 pKa = 11.84SSDD520 pKa = 3.67TADD523 pKa = 3.12MPLNGTATPGSRR535 pKa = 11.84YY536 pKa = 10.06AYY538 pKa = 9.86AAGGRR543 pKa = 11.84DD544 pKa = 3.69VLRR547 pKa = 11.84VIGVTPDD554 pKa = 2.91VRR556 pKa = 11.84KK557 pKa = 9.93SVLPITVDD565 pKa = 3.37PSGEE569 pKa = 3.76ATYY572 pKa = 10.49TLTYY576 pKa = 9.71SANGTGAVAPTVDD589 pKa = 5.23DD590 pKa = 3.95YY591 pKa = 12.31VLVDD595 pKa = 4.0TLPAGATYY603 pKa = 10.82VGGSASPEE611 pKa = 3.86PAVTTNGSGQQVLTWTLDD629 pKa = 3.51GVATNAEE636 pKa = 4.18HH637 pKa = 6.16SLSYY641 pKa = 10.46VVRR644 pKa = 11.84FPDD647 pKa = 4.15DD648 pKa = 3.49LQGGDD653 pKa = 3.21RR654 pKa = 11.84LVNTVSAAVEE664 pKa = 4.5GVTSDD669 pKa = 3.81DD670 pKa = 3.29ATASVVVSDD679 pKa = 4.58SGLTKK684 pKa = 10.02IIKK687 pKa = 8.82TADD690 pKa = 3.09QAFIPNLAGDD700 pKa = 4.34GVGTGSWTVLVRR712 pKa = 11.84SQDD715 pKa = 3.85PIAQSFVDD723 pKa = 4.98VIDD726 pKa = 3.49ILPYY730 pKa = 10.33VGDD733 pKa = 3.75GRR735 pKa = 11.84GTAFSGSYY743 pKa = 10.68LLDD746 pKa = 3.98GPVTSSIGGTVYY758 pKa = 9.49YY759 pKa = 7.49TTADD763 pKa = 3.77PTTLTDD769 pKa = 5.21DD770 pKa = 4.48PADD773 pKa = 4.2PSNGSAGSVAGNTVGWTTEE792 pKa = 3.93YY793 pKa = 10.77TPDD796 pKa = 3.1ATAVRR801 pKa = 11.84VITGTLAPRR810 pKa = 11.84GEE812 pKa = 4.36LTISVPVVTDD822 pKa = 3.21GMVGGDD828 pKa = 3.36TLVNRR833 pKa = 11.84SQGRR837 pKa = 11.84AEE839 pKa = 3.9NTRR842 pKa = 11.84LVMRR846 pKa = 11.84TSAPTSIANYY856 pKa = 10.02YY857 pKa = 9.47SASLKK862 pKa = 10.66KK863 pKa = 10.44YY864 pKa = 9.53VQDD867 pKa = 5.32ADD869 pKa = 4.43GEE871 pKa = 4.37WRR873 pKa = 11.84DD874 pKa = 3.72ANDD877 pKa = 3.96PLDD880 pKa = 3.91YY881 pKa = 10.67PQFRR885 pKa = 11.84VGDD888 pKa = 3.75TVRR891 pKa = 11.84YY892 pKa = 9.3RR893 pKa = 11.84IVVEE897 pKa = 3.83NTGQGTLTNVVVSDD911 pKa = 4.21DD912 pKa = 4.09LQPEE916 pKa = 4.52LGSFTIEE923 pKa = 3.76SLAPGEE929 pKa = 4.43VEE931 pKa = 3.81SHH933 pKa = 5.59EE934 pKa = 4.23FEE936 pKa = 4.35IVLSEE941 pKa = 4.11PVDD944 pKa = 3.67DD945 pKa = 4.79AVVNTACADD954 pKa = 3.47ADD956 pKa = 3.81VPEE959 pKa = 5.03DD960 pKa = 3.61SGVEE964 pKa = 3.82PTINCDD970 pKa = 3.08PAGFTVVGTPTHH982 pKa = 5.34TKK984 pKa = 9.57EE985 pKa = 4.21IVSATPVGDD994 pKa = 3.73GRR996 pKa = 11.84WEE998 pKa = 3.86VVYY1001 pKa = 10.24EE1002 pKa = 4.08VVVSNTQTPSTTYY1015 pKa = 10.84SLTDD1019 pKa = 3.4EE1020 pKa = 5.01LHH1022 pKa = 6.03FTDD1025 pKa = 3.54QATIVSATVTQAPDD1039 pKa = 3.58GVVLADD1045 pKa = 4.12PAWDD1049 pKa = 3.67GQEE1052 pKa = 4.05NLTIASDD1059 pKa = 3.94VPLAGNDD1066 pKa = 3.44DD1067 pKa = 4.44DD1068 pKa = 6.15GYY1070 pKa = 9.77EE1071 pKa = 3.86PHH1073 pKa = 6.83VYY1075 pKa = 9.82RR1076 pKa = 11.84VTVLADD1082 pKa = 3.21VPLQLDD1088 pKa = 4.16GAGSGADD1095 pKa = 4.74DD1096 pKa = 3.83PTACPAEE1103 pKa = 4.67GSDD1106 pKa = 4.92ADD1108 pKa = 3.46QGFNNTSGLTGPDD1121 pKa = 3.52GEE1123 pKa = 4.59TEE1125 pKa = 4.02EE1126 pKa = 5.26DD1127 pKa = 3.73QACAPIPSIDD1137 pKa = 3.44VEE1139 pKa = 4.34KK1140 pKa = 10.55RR1141 pKa = 11.84VAAGPTPNGDD1151 pKa = 3.68GTWTVTYY1158 pKa = 10.57DD1159 pKa = 3.59VVATNSGTGDD1169 pKa = 3.25GVYY1172 pKa = 10.42EE1173 pKa = 4.15VTDD1176 pKa = 3.49RR1177 pKa = 11.84MTADD1181 pKa = 3.33GDD1183 pKa = 4.17LEE1185 pKa = 4.6VVSGAVTSAPDD1196 pKa = 3.62GVTPSATWTGLGAEE1210 pKa = 4.51GAPEE1214 pKa = 3.87NVIASAVTLPAGTSHH1229 pKa = 7.15AYY1231 pKa = 8.88EE1232 pKa = 4.18VQVVLTIAPGTEE1244 pKa = 3.95GAPVVTPCSAEE1255 pKa = 3.86PGGSSGGLSNAAGVEE1270 pKa = 4.35HH1271 pKa = 7.47NDD1273 pKa = 3.45LTDD1276 pKa = 3.28TAEE1279 pKa = 4.23ACVTLAAITVEE1290 pKa = 4.4KK1291 pKa = 10.24SISAGPTPNGDD1302 pKa = 3.71GTWTVVYY1309 pKa = 10.37DD1310 pKa = 4.06VVAEE1314 pKa = 4.16NVGAAAGEE1322 pKa = 3.96YY1323 pKa = 10.18DD1324 pKa = 3.48VYY1326 pKa = 11.69DD1327 pKa = 3.63RR1328 pKa = 11.84LRR1330 pKa = 11.84YY1331 pKa = 9.77GDD1333 pKa = 4.22GIEE1336 pKa = 4.2IVDD1339 pKa = 3.89AAVTSGPDD1347 pKa = 3.39GVTTNAAWTGLGEE1360 pKa = 4.78PDD1362 pKa = 3.09TAAEE1366 pKa = 4.15NLVAADD1372 pKa = 3.7VEE1374 pKa = 4.48LAAGGLHH1381 pKa = 5.92TFQVEE1386 pKa = 4.13VTVRR1390 pKa = 11.84MDD1392 pKa = 3.22EE1393 pKa = 4.03ATIDD1397 pKa = 3.94PVALRR1402 pKa = 11.84CPPPGSGASGGLANSTSLDD1421 pKa = 3.39HH1422 pKa = 6.64NGIVAEE1428 pKa = 4.69DD1429 pKa = 4.23EE1430 pKa = 4.67VCASLPLIGVEE1441 pKa = 4.39KK1442 pKa = 10.27SISAGPVPQGDD1453 pKa = 4.2GTWAITYY1460 pKa = 10.48DD1461 pKa = 4.51LVATNAGEE1469 pKa = 3.96AAGDD1473 pKa = 3.59YY1474 pKa = 11.2DD1475 pKa = 5.31LVDD1478 pKa = 3.49RR1479 pKa = 11.84LQYY1482 pKa = 10.91GEE1484 pKa = 4.3GVVVEE1489 pKa = 4.48SAAVVSAPDD1498 pKa = 3.83GVATEE1503 pKa = 4.8AGWTGQGAAGSTEE1516 pKa = 3.95NVVAAGVTLGAGAAHH1531 pKa = 7.38TYY1533 pKa = 8.7QVQVVVSLDD1542 pKa = 3.48RR1543 pKa = 11.84AVVTPSTLACPEE1555 pKa = 4.54PGSGEE1560 pKa = 4.16GGGLANATEE1569 pKa = 4.29LTHH1572 pKa = 7.49NGEE1575 pKa = 4.31TQGDD1579 pKa = 4.21DD1580 pKa = 3.41VCAEE1584 pKa = 4.43LPLIDD1589 pKa = 3.88IAKK1592 pKa = 8.81SLSGAVTPVEE1602 pKa = 4.32GEE1604 pKa = 3.9PGVYY1608 pKa = 10.07DD1609 pKa = 3.68AVYY1612 pKa = 10.34EE1613 pKa = 4.17ITVTNAGPGDD1623 pKa = 3.97GLYY1626 pKa = 11.08DD1627 pKa = 3.93LTDD1630 pKa = 3.39EE1631 pKa = 4.6LAPGEE1636 pKa = 4.23GVEE1639 pKa = 4.15VLGVQDD1645 pKa = 4.0VVTDD1649 pKa = 3.75APEE1652 pKa = 3.94PVGLNPGFDD1661 pKa = 3.71GTSDD1665 pKa = 3.6TLVVAGQPIAGATTAPVVHH1684 pKa = 7.1TYY1686 pKa = 9.23TVTVRR1691 pKa = 11.84YY1692 pKa = 9.04AADD1695 pKa = 3.72LSDD1698 pKa = 4.69VEE1700 pKa = 5.49IPDD1703 pKa = 3.63TDD1705 pKa = 3.28ACTTGDD1711 pKa = 3.45GSTVAGGLANTATVGWNGLDD1731 pKa = 3.76GTDD1734 pKa = 4.14DD1735 pKa = 3.54ACVRR1739 pKa = 11.84PGKK1742 pKa = 8.38PTLDD1746 pKa = 3.44KK1747 pKa = 11.02ALVSAAPAGDD1757 pKa = 3.84GTWEE1761 pKa = 4.04VVYY1764 pKa = 10.66DD1765 pKa = 3.7LTVGNVGGEE1774 pKa = 3.86ATTYY1778 pKa = 11.33DD1779 pKa = 4.64LDD1781 pKa = 5.91DD1782 pKa = 4.55EE1783 pKa = 5.49LLFAPVVTVDD1793 pKa = 3.56SVAVTGPEE1801 pKa = 3.91GVTVNAGFDD1810 pKa = 3.34GATDD1814 pKa = 3.43QRR1816 pKa = 11.84IATGVSIAGMDD1827 pKa = 3.73DD1828 pKa = 3.11EE1829 pKa = 6.21GYY1831 pKa = 10.42APHH1834 pKa = 7.05VYY1836 pKa = 9.69RR1837 pKa = 11.84VTVVAHH1843 pKa = 5.84VPLSFAADD1851 pKa = 3.73DD1852 pKa = 3.8VDD1854 pKa = 4.83GDD1856 pKa = 4.33GTGSPACTVPAGSNTLQQGLNNAATLTDD1884 pKa = 3.88EE1885 pKa = 4.55TGGTQTDD1892 pKa = 3.89TDD1894 pKa = 4.43CAPLPSIDD1902 pKa = 3.66VTKK1905 pKa = 10.33STDD1908 pKa = 3.27GDD1910 pKa = 4.0PVQGDD1915 pKa = 3.61DD1916 pKa = 4.03GSWTVRR1922 pKa = 11.84YY1923 pKa = 9.43AVTVTNDD1930 pKa = 2.98GGSAGAYY1937 pKa = 10.1DD1938 pKa = 3.59LTDD1941 pKa = 3.5RR1942 pKa = 11.84LRR1944 pKa = 11.84YY1945 pKa = 8.97GAGITVRR1952 pKa = 11.84SATVTATPEE1961 pKa = 4.09GVTASATWTGQGAEE1975 pKa = 3.8GDD1977 pKa = 3.81AANVVAADD1985 pKa = 3.42VDD1987 pKa = 4.35LAPGATHH1994 pKa = 6.66VYY1996 pKa = 9.41RR1997 pKa = 11.84VEE1999 pKa = 3.96VVAAVDD2005 pKa = 3.76PGRR2008 pKa = 11.84ADD2010 pKa = 3.07ATTFTCPAPGSGEE2023 pKa = 3.81AGGFANTAGVGHH2035 pKa = 7.01NDD2037 pKa = 3.09LTDD2040 pKa = 3.84AADD2043 pKa = 3.48ACAAPPEE2050 pKa = 4.38PANPGTPGKK2059 pKa = 9.67PGGSLATTGATLAWVAGGAALLLLLGMLALLAARR2093 pKa = 11.84RR2094 pKa = 11.84RR2095 pKa = 11.84RR2096 pKa = 11.84LTSS2099 pKa = 2.99
Molecular weight: 213.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7CNR9|A0A1X7CNR9_CELCE Inositol-1-monophosphatase OS=Cellulosimicrobium cellulans J1 OX=903523 GN=SAMN02744115_00806 PE=3 SV=1
MM1 pKa = 7.63AGLRR5 pKa = 11.84PAVEE9 pKa = 4.22PVAVPARR16 pKa = 11.84AGARR20 pKa = 11.84RR21 pKa = 11.84VPTPVAPATVVARR34 pKa = 11.84PARR37 pKa = 11.84STTSVRR43 pKa = 11.84RR44 pKa = 11.84VPVEE48 pKa = 3.84PPAAALPRR56 pKa = 11.84ARR58 pKa = 11.84GPLRR62 pKa = 11.84VAPLTSVAVALARR75 pKa = 11.84PPVVPTRR82 pKa = 11.84GTVVTARR89 pKa = 11.84GTVAATRR96 pKa = 11.84TLVATGVPAAVLAARR111 pKa = 11.84AVGTVLAARR120 pKa = 11.84TAPGSIPTGAVTSVAGAVTTVRR142 pKa = 11.84VRR144 pKa = 11.84ATAVGSTVVATVALRR159 pKa = 11.84TAALGTVRR167 pKa = 11.84TRR169 pKa = 11.84ASTVGSVPVAALRR182 pKa = 11.84APAPVARR189 pKa = 11.84AALVTAAATVTRR201 pKa = 11.84RR202 pKa = 11.84VTAGRR207 pKa = 11.84PAMSPATLVAGASSTVLVASTAAVVARR234 pKa = 11.84PRR236 pKa = 11.84TVRR239 pKa = 11.84LGLVGAVVHH248 pKa = 6.23EE249 pKa = 4.92GSLSFWSLRR258 pKa = 11.84RR259 pKa = 11.84PSSHH263 pKa = 6.85ASVLGHH269 pKa = 6.53AAGRR273 pKa = 11.84LL274 pKa = 3.55
MM1 pKa = 7.63AGLRR5 pKa = 11.84PAVEE9 pKa = 4.22PVAVPARR16 pKa = 11.84AGARR20 pKa = 11.84RR21 pKa = 11.84VPTPVAPATVVARR34 pKa = 11.84PARR37 pKa = 11.84STTSVRR43 pKa = 11.84RR44 pKa = 11.84VPVEE48 pKa = 3.84PPAAALPRR56 pKa = 11.84ARR58 pKa = 11.84GPLRR62 pKa = 11.84VAPLTSVAVALARR75 pKa = 11.84PPVVPTRR82 pKa = 11.84GTVVTARR89 pKa = 11.84GTVAATRR96 pKa = 11.84TLVATGVPAAVLAARR111 pKa = 11.84AVGTVLAARR120 pKa = 11.84TAPGSIPTGAVTSVAGAVTTVRR142 pKa = 11.84VRR144 pKa = 11.84ATAVGSTVVATVALRR159 pKa = 11.84TAALGTVRR167 pKa = 11.84TRR169 pKa = 11.84ASTVGSVPVAALRR182 pKa = 11.84APAPVARR189 pKa = 11.84AALVTAAATVTRR201 pKa = 11.84RR202 pKa = 11.84VTAGRR207 pKa = 11.84PAMSPATLVAGASSTVLVASTAAVVARR234 pKa = 11.84PRR236 pKa = 11.84TVRR239 pKa = 11.84LGLVGAVVHH248 pKa = 6.23EE249 pKa = 4.92GSLSFWSLRR258 pKa = 11.84RR259 pKa = 11.84PSSHH263 pKa = 6.85ASVLGHH269 pKa = 6.53AAGRR273 pKa = 11.84LL274 pKa = 3.55
Molecular weight: 26.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1281283 |
27 |
2099 |
342.3 |
36.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.844 ± 0.06 | 0.512 ± 0.01 |
6.72 ± 0.036 | 5.42 ± 0.03 |
2.516 ± 0.023 | 9.564 ± 0.036 |
2.02 ± 0.019 | 2.536 ± 0.026 |
1.343 ± 0.023 | 9.969 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.406 ± 0.014 | 1.473 ± 0.022 |
6.25 ± 0.035 | 2.433 ± 0.021 |
7.985 ± 0.05 | 4.761 ± 0.023 |
6.398 ± 0.043 | 10.445 ± 0.041 |
1.544 ± 0.016 | 1.861 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
