
Eggplant mottled dwarf nucleorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
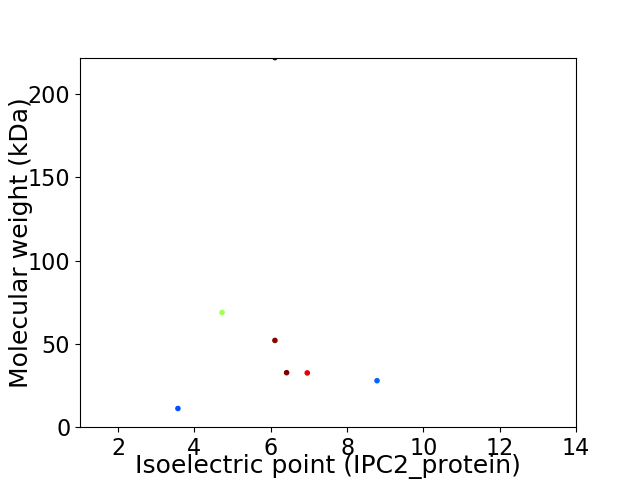
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140KP54|A0A140KP54_9RHAB Phosphoprotein OS=Eggplant mottled dwarf nucleorhabdovirus OX=488317 GN=P PE=4 SV=1
MM1 pKa = 7.88SEE3 pKa = 4.0TTTNRR8 pKa = 11.84PITPDD13 pKa = 3.26EE14 pKa = 4.59ANDD17 pKa = 3.73DD18 pKa = 4.19VFVEE22 pKa = 4.39MCRR25 pKa = 11.84MVDD28 pKa = 3.33NLLSQSEE35 pKa = 4.7TNKK38 pKa = 10.13HH39 pKa = 6.38DD40 pKa = 4.22ANQDD44 pKa = 3.83SIPITIEE51 pKa = 3.8EE52 pKa = 4.31EE53 pKa = 4.01TSDD56 pKa = 4.96AEE58 pKa = 5.28SSDD61 pKa = 2.58SWTYY65 pKa = 11.63YY66 pKa = 11.08DD67 pKa = 3.74FTDD70 pKa = 4.22GRR72 pKa = 11.84DD73 pKa = 3.36YY74 pKa = 11.47YY75 pKa = 11.67YY76 pKa = 10.92DD77 pKa = 5.27DD78 pKa = 4.68NDD80 pKa = 3.88SDD82 pKa = 3.83IYY84 pKa = 11.43EE85 pKa = 4.53LMAEE89 pKa = 4.52CNHH92 pKa = 6.34GEE94 pKa = 3.94WGNN97 pKa = 3.42
MM1 pKa = 7.88SEE3 pKa = 4.0TTTNRR8 pKa = 11.84PITPDD13 pKa = 3.26EE14 pKa = 4.59ANDD17 pKa = 3.73DD18 pKa = 4.19VFVEE22 pKa = 4.39MCRR25 pKa = 11.84MVDD28 pKa = 3.33NLLSQSEE35 pKa = 4.7TNKK38 pKa = 10.13HH39 pKa = 6.38DD40 pKa = 4.22ANQDD44 pKa = 3.83SIPITIEE51 pKa = 3.8EE52 pKa = 4.31EE53 pKa = 4.01TSDD56 pKa = 4.96AEE58 pKa = 5.28SSDD61 pKa = 2.58SWTYY65 pKa = 11.63YY66 pKa = 11.08DD67 pKa = 3.74FTDD70 pKa = 4.22GRR72 pKa = 11.84DD73 pKa = 3.36YY74 pKa = 11.47YY75 pKa = 11.67YY76 pKa = 10.92DD77 pKa = 5.27DD78 pKa = 4.68NDD80 pKa = 3.88SDD82 pKa = 3.83IYY84 pKa = 11.43EE85 pKa = 4.53LMAEE89 pKa = 4.52CNHH92 pKa = 6.34GEE94 pKa = 3.94WGNN97 pKa = 3.42
Molecular weight: 11.22 kDa
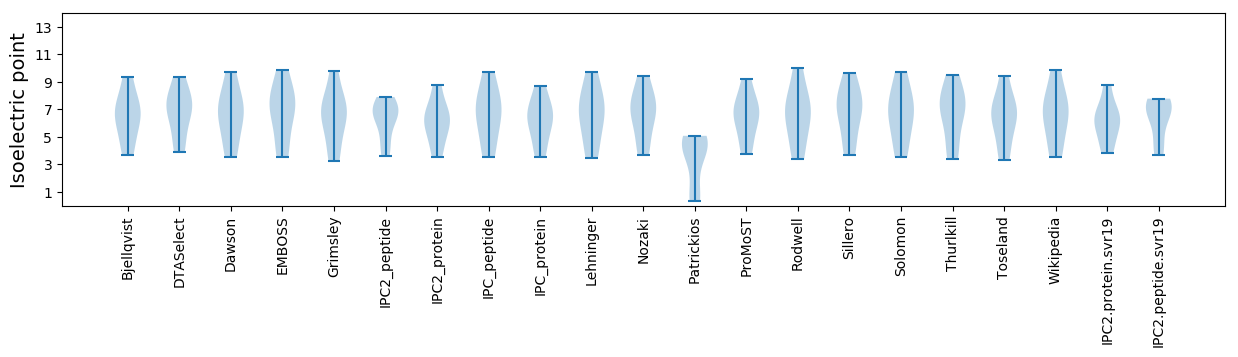
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140KP58|A0A140KP58_9RHAB Large structural protein OS=Eggplant mottled dwarf nucleorhabdovirus OX=488317 GN=G PE=4 SV=1
MM1 pKa = 7.45INVIGRR7 pKa = 11.84SSASVQGIYY16 pKa = 10.28DD17 pKa = 3.56VEE19 pKa = 4.17KK20 pKa = 11.07DD21 pKa = 3.63KK22 pKa = 11.73VSLLSTVVGQPISLNYY38 pKa = 9.84IITIHH43 pKa = 5.97MYY45 pKa = 8.57EE46 pKa = 4.6AEE48 pKa = 4.09MKK50 pKa = 10.41KK51 pKa = 10.43AFEE54 pKa = 4.3EE55 pKa = 5.19GEE57 pKa = 4.18LHH59 pKa = 5.76YY60 pKa = 10.86HH61 pKa = 7.52DD62 pKa = 4.98IFDD65 pKa = 4.5MIEE68 pKa = 4.03SALEE72 pKa = 4.07LPSTRR77 pKa = 11.84PDD79 pKa = 3.16AVKK82 pKa = 10.37RR83 pKa = 11.84SPPNMTARR91 pKa = 11.84VAKK94 pKa = 10.43NILQICRR101 pKa = 11.84IHH103 pKa = 7.53AIHH106 pKa = 6.74TNAKK110 pKa = 7.93MVFMNTSTDD119 pKa = 3.22HH120 pKa = 6.91SNEE123 pKa = 3.96TPTLVIKK130 pKa = 10.62EE131 pKa = 4.27GEE133 pKa = 4.32TLVSQNYY140 pKa = 8.97DD141 pKa = 3.24SHH143 pKa = 7.93VVEE146 pKa = 5.28IVNQAGDD153 pKa = 3.66NPLSGSYY160 pKa = 10.41HH161 pKa = 5.65LTLEE165 pKa = 4.19KK166 pKa = 10.35PYY168 pKa = 10.09ISSDD172 pKa = 3.16SRR174 pKa = 11.84IVGHH178 pKa = 6.93LSFAAYY184 pKa = 9.66VRR186 pKa = 11.84APPRR190 pKa = 11.84GQAATRR196 pKa = 11.84TRR198 pKa = 11.84LNIRR202 pKa = 11.84VLASKK207 pKa = 8.29YY208 pKa = 8.15QRR210 pKa = 11.84NASQKK215 pKa = 9.4VNSLFRR221 pKa = 11.84KK222 pKa = 8.87SIKK225 pKa = 10.27RR226 pKa = 11.84KK227 pKa = 10.19GDD229 pKa = 4.24DD230 pKa = 3.14ISKK233 pKa = 9.89KK234 pKa = 8.3PHH236 pKa = 6.23SSGIMDD242 pKa = 4.57AVAKK246 pKa = 10.03ILKK249 pKa = 10.29SSS251 pKa = 3.27
MM1 pKa = 7.45INVIGRR7 pKa = 11.84SSASVQGIYY16 pKa = 10.28DD17 pKa = 3.56VEE19 pKa = 4.17KK20 pKa = 11.07DD21 pKa = 3.63KK22 pKa = 11.73VSLLSTVVGQPISLNYY38 pKa = 9.84IITIHH43 pKa = 5.97MYY45 pKa = 8.57EE46 pKa = 4.6AEE48 pKa = 4.09MKK50 pKa = 10.41KK51 pKa = 10.43AFEE54 pKa = 4.3EE55 pKa = 5.19GEE57 pKa = 4.18LHH59 pKa = 5.76YY60 pKa = 10.86HH61 pKa = 7.52DD62 pKa = 4.98IFDD65 pKa = 4.5MIEE68 pKa = 4.03SALEE72 pKa = 4.07LPSTRR77 pKa = 11.84PDD79 pKa = 3.16AVKK82 pKa = 10.37RR83 pKa = 11.84SPPNMTARR91 pKa = 11.84VAKK94 pKa = 10.43NILQICRR101 pKa = 11.84IHH103 pKa = 7.53AIHH106 pKa = 6.74TNAKK110 pKa = 7.93MVFMNTSTDD119 pKa = 3.22HH120 pKa = 6.91SNEE123 pKa = 3.96TPTLVIKK130 pKa = 10.62EE131 pKa = 4.27GEE133 pKa = 4.32TLVSQNYY140 pKa = 8.97DD141 pKa = 3.24SHH143 pKa = 7.93VVEE146 pKa = 5.28IVNQAGDD153 pKa = 3.66NPLSGSYY160 pKa = 10.41HH161 pKa = 5.65LTLEE165 pKa = 4.19KK166 pKa = 10.35PYY168 pKa = 10.09ISSDD172 pKa = 3.16SRR174 pKa = 11.84IVGHH178 pKa = 6.93LSFAAYY184 pKa = 9.66VRR186 pKa = 11.84APPRR190 pKa = 11.84GQAATRR196 pKa = 11.84TRR198 pKa = 11.84LNIRR202 pKa = 11.84VLASKK207 pKa = 8.29YY208 pKa = 8.15QRR210 pKa = 11.84NASQKK215 pKa = 9.4VNSLFRR221 pKa = 11.84KK222 pKa = 8.87SIKK225 pKa = 10.27RR226 pKa = 11.84KK227 pKa = 10.19GDD229 pKa = 4.24DD230 pKa = 3.14ISKK233 pKa = 9.89KK234 pKa = 8.3PHH236 pKa = 6.23SSGIMDD242 pKa = 4.57AVAKK246 pKa = 10.03ILKK249 pKa = 10.29SSS251 pKa = 3.27
Molecular weight: 27.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3966 |
97 |
1946 |
566.6 |
63.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.018 ± 1.022 | 1.664 ± 0.45 |
6.43 ± 0.365 | 5.522 ± 0.465 |
3.404 ± 0.355 | 5.673 ± 0.297 |
2.37 ± 0.436 | 7.892 ± 0.286 |
5.547 ± 0.626 | 8.447 ± 0.856 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.833 ± 0.352 | 4.639 ± 0.417 |
4.791 ± 0.6 | 3.127 ± 0.283 |
5.169 ± 0.811 | 9.178 ± 0.898 |
6.051 ± 0.467 | 6.203 ± 0.286 |
1.16 ± 0.302 | 3.883 ± 0.413 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
