
Gaetbulibacter sp. 4G1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gaetbulibacter; unclassified Gaetbulibacter
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
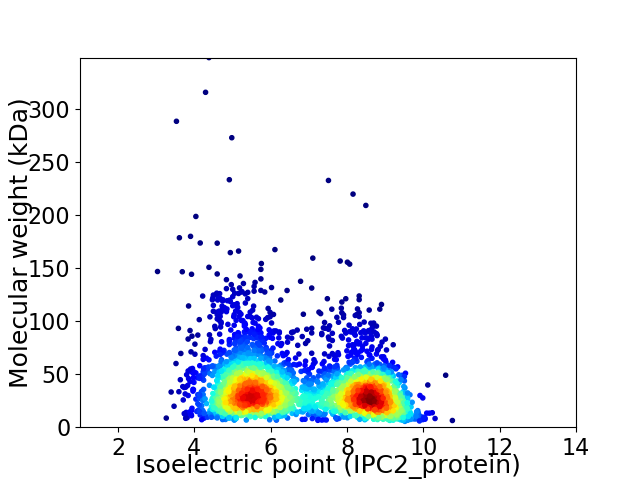
Virtual 2D-PAGE plot for 3344 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G5GH64|A0A2G5GH64_9FLAO Uncharacterized protein OS=Gaetbulibacter sp. 4G1 OX=1891971 GN=BFR04_01600 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.55KK3 pKa = 10.42NILLLCIALLCVHH16 pKa = 6.46VLNAQCGPGEE26 pKa = 4.44DD27 pKa = 3.11TTAPVFGNAGDD38 pKa = 3.8GTMANPFRR46 pKa = 11.84NLLLSTVGGVPSGTYY61 pKa = 9.44YY62 pKa = 11.03FNFNGSTFQGALDD75 pKa = 4.11NDD77 pKa = 4.02TDD79 pKa = 3.91GGGWLMVLNYY89 pKa = 10.17VHH91 pKa = 7.36LAGDD95 pKa = 3.78NTDD98 pKa = 3.23LTIRR102 pKa = 11.84NTDD105 pKa = 3.47LPLLGSSTLGDD116 pKa = 3.8NEE118 pKa = 5.08AGTANWGHH126 pKa = 6.5MGNALAAAIDD136 pKa = 3.85FEE138 pKa = 4.79EE139 pKa = 3.83IRR141 pKa = 11.84FYY143 pKa = 11.63GEE145 pKa = 4.07TTGHH149 pKa = 4.66TRR151 pKa = 11.84IINFKK156 pKa = 8.74TPYY159 pKa = 9.94ISGLNYY165 pKa = 10.35LKK167 pKa = 10.16TGLGHH172 pKa = 7.11FGGINNPSNFTTLTGHH188 pKa = 6.32TANIPALAGNFFASHH203 pKa = 6.77GDD205 pKa = 3.34NALTEE210 pKa = 4.62FPFFSSGQNHH220 pKa = 5.55WGIRR224 pKa = 11.84GSGNRR229 pKa = 11.84WEE231 pKa = 4.8VDD233 pKa = 3.59DD234 pKa = 5.69APTAASPVNSQSTIHH249 pKa = 5.99RR250 pKa = 11.84VWVRR254 pKa = 11.84GDD256 pKa = 3.77LSPAGTPEE264 pKa = 4.18VSVTLDD270 pKa = 2.97NTGNVTVNPADD281 pKa = 3.65FGYY284 pKa = 8.86TVMDD288 pKa = 3.83NCSLEE293 pKa = 4.1ANINISLSQTDD304 pKa = 4.08FACIDD309 pKa = 3.32VGANIIQFTVTDD321 pKa = 3.87EE322 pKa = 4.37QNNSTVIDD330 pKa = 3.61VTVTVLEE337 pKa = 4.35SPPVITTPSGPFQRR351 pKa = 11.84ISLDD355 pKa = 3.65SNGEE359 pKa = 3.96LTLDD363 pKa = 3.92LATLNVTATDD373 pKa = 3.7DD374 pKa = 3.72CGIASTVISPATLDD388 pKa = 3.79CSDD391 pKa = 4.9AGFKK395 pKa = 10.54NATITVTDD403 pKa = 3.63VNGNASNSSMVFILIDD419 pKa = 3.32QVPPVIQCVPSYY431 pKa = 9.58TAEE434 pKa = 4.17LDD436 pKa = 3.59GTGSVTLDD444 pKa = 3.55PNSLLTSFTDD454 pKa = 3.28NCQTAGVSLDD464 pKa = 3.42KK465 pKa = 11.02TVFTCADD472 pKa = 3.08IGDD475 pKa = 4.08NLVTVTATDD484 pKa = 3.64GDD486 pKa = 4.67GNSTTCTTTVTITIPSCPGNLTLQAEE512 pKa = 4.35PNACGVTYY520 pKa = 10.4NYY522 pKa = 9.82PCASNVTAGPPSGTFLAVGTTTTFAYY548 pKa = 7.87DD549 pKa = 3.36TLDD552 pKa = 3.52NTGGTVTCSYY562 pKa = 11.24DD563 pKa = 3.27VTVVDD568 pKa = 4.3EE569 pKa = 4.75EE570 pKa = 5.19GPTFATQNITLDD582 pKa = 3.42VTMDD586 pKa = 3.46GTVSIVAEE594 pKa = 4.38DD595 pKa = 4.1LLGLDD600 pKa = 5.1LLASDD605 pKa = 4.18YY606 pKa = 11.15TVDD609 pKa = 3.46QTGTFNRR616 pKa = 11.84EE617 pKa = 4.27DD618 pKa = 3.19ISTTGTEE625 pKa = 3.53ISLFDD630 pKa = 4.21DD631 pKa = 3.9QVSSALPIGFEE642 pKa = 3.92FAFYY646 pKa = 10.46GALYY650 pKa = 10.06NDD652 pKa = 5.25FYY654 pKa = 10.94ISSNGFITFSGNSDD668 pKa = 3.83DD669 pKa = 4.73GCCTGQTLPDD679 pKa = 3.3VTEE682 pKa = 4.29PNNLIAFDD690 pKa = 3.81WDD692 pKa = 4.05DD693 pKa = 4.17YY694 pKa = 11.6DD695 pKa = 4.38PEE697 pKa = 5.63SGGTIRR703 pKa = 11.84YY704 pKa = 5.8QTIGTAPNRR713 pKa = 11.84ILIMDD718 pKa = 4.45FDD720 pKa = 4.69AVLHH724 pKa = 6.64IDD726 pKa = 3.77YY727 pKa = 10.94NDD729 pKa = 3.41NTDD732 pKa = 3.04ATTTQVKK739 pKa = 10.15LFEE742 pKa = 4.31GTNRR746 pKa = 11.84IEE748 pKa = 3.75IHH750 pKa = 5.91ATNIPDD756 pKa = 4.23LSNTKK761 pKa = 8.31TQGLEE766 pKa = 4.23NIDD769 pKa = 3.29GTAAIIVPGRR779 pKa = 11.84NAATWSASNDD789 pKa = 3.02VVAFVPVTGGVADD802 pKa = 3.64SCGVDD807 pKa = 3.2TFVASQTTFDD817 pKa = 4.5CSNLGANTVTLTATDD832 pKa = 3.54TNGNITEE839 pKa = 4.77KK840 pKa = 10.49IAIVTVTDD848 pKa = 4.22ANNFCPDD855 pKa = 3.11ILVSPKK861 pKa = 10.36VFLQGAALNPNVGEE875 pKa = 4.12EE876 pKa = 3.94TLMRR880 pKa = 11.84DD881 pKa = 3.98DD882 pKa = 5.64LRR884 pKa = 11.84IAGQIPTTSPYY895 pKa = 10.55TDD897 pKa = 4.25GLTCDD902 pKa = 3.55ASVFTVTGANAIVDD916 pKa = 4.23WVWLEE921 pKa = 3.69LRR923 pKa = 11.84DD924 pKa = 4.79PITSTTIVSSRR935 pKa = 11.84SALLQRR941 pKa = 11.84DD942 pKa = 3.61GDD944 pKa = 3.91IVDD947 pKa = 3.6IDD949 pKa = 4.06GVSTSVIFSEE959 pKa = 5.13VPGAYY964 pKa = 9.97YY965 pKa = 10.65LVVTHH970 pKa = 7.08RR971 pKa = 11.84NHH973 pKa = 6.83LGIRR977 pKa = 11.84SANTYY982 pKa = 9.88NLSSTTTTINLSTSSNSVAGGTNAIIDD1009 pKa = 3.99LGNGKK1014 pKa = 10.09FGLIGGDD1021 pKa = 3.66FDD1023 pKa = 6.99ANGQVQNTDD1032 pKa = 3.04LSGVRR1037 pKa = 11.84PVLGQSQYY1045 pKa = 11.52SAGDD1049 pKa = 3.48LDD1051 pKa = 4.62MNSQVQNTDD1060 pKa = 2.38INNLLNPNTGKK1071 pKa = 10.77GKK1073 pKa = 9.98QFAKK1077 pKa = 10.41QNLRR1081 pKa = 11.84AKK1083 pKa = 10.12LKK1085 pKa = 10.52KK1086 pKa = 10.36
MM1 pKa = 7.66KK2 pKa = 10.55KK3 pKa = 10.42NILLLCIALLCVHH16 pKa = 6.46VLNAQCGPGEE26 pKa = 4.44DD27 pKa = 3.11TTAPVFGNAGDD38 pKa = 3.8GTMANPFRR46 pKa = 11.84NLLLSTVGGVPSGTYY61 pKa = 9.44YY62 pKa = 11.03FNFNGSTFQGALDD75 pKa = 4.11NDD77 pKa = 4.02TDD79 pKa = 3.91GGGWLMVLNYY89 pKa = 10.17VHH91 pKa = 7.36LAGDD95 pKa = 3.78NTDD98 pKa = 3.23LTIRR102 pKa = 11.84NTDD105 pKa = 3.47LPLLGSSTLGDD116 pKa = 3.8NEE118 pKa = 5.08AGTANWGHH126 pKa = 6.5MGNALAAAIDD136 pKa = 3.85FEE138 pKa = 4.79EE139 pKa = 3.83IRR141 pKa = 11.84FYY143 pKa = 11.63GEE145 pKa = 4.07TTGHH149 pKa = 4.66TRR151 pKa = 11.84IINFKK156 pKa = 8.74TPYY159 pKa = 9.94ISGLNYY165 pKa = 10.35LKK167 pKa = 10.16TGLGHH172 pKa = 7.11FGGINNPSNFTTLTGHH188 pKa = 6.32TANIPALAGNFFASHH203 pKa = 6.77GDD205 pKa = 3.34NALTEE210 pKa = 4.62FPFFSSGQNHH220 pKa = 5.55WGIRR224 pKa = 11.84GSGNRR229 pKa = 11.84WEE231 pKa = 4.8VDD233 pKa = 3.59DD234 pKa = 5.69APTAASPVNSQSTIHH249 pKa = 5.99RR250 pKa = 11.84VWVRR254 pKa = 11.84GDD256 pKa = 3.77LSPAGTPEE264 pKa = 4.18VSVTLDD270 pKa = 2.97NTGNVTVNPADD281 pKa = 3.65FGYY284 pKa = 8.86TVMDD288 pKa = 3.83NCSLEE293 pKa = 4.1ANINISLSQTDD304 pKa = 4.08FACIDD309 pKa = 3.32VGANIIQFTVTDD321 pKa = 3.87EE322 pKa = 4.37QNNSTVIDD330 pKa = 3.61VTVTVLEE337 pKa = 4.35SPPVITTPSGPFQRR351 pKa = 11.84ISLDD355 pKa = 3.65SNGEE359 pKa = 3.96LTLDD363 pKa = 3.92LATLNVTATDD373 pKa = 3.7DD374 pKa = 3.72CGIASTVISPATLDD388 pKa = 3.79CSDD391 pKa = 4.9AGFKK395 pKa = 10.54NATITVTDD403 pKa = 3.63VNGNASNSSMVFILIDD419 pKa = 3.32QVPPVIQCVPSYY431 pKa = 9.58TAEE434 pKa = 4.17LDD436 pKa = 3.59GTGSVTLDD444 pKa = 3.55PNSLLTSFTDD454 pKa = 3.28NCQTAGVSLDD464 pKa = 3.42KK465 pKa = 11.02TVFTCADD472 pKa = 3.08IGDD475 pKa = 4.08NLVTVTATDD484 pKa = 3.64GDD486 pKa = 4.67GNSTTCTTTVTITIPSCPGNLTLQAEE512 pKa = 4.35PNACGVTYY520 pKa = 10.4NYY522 pKa = 9.82PCASNVTAGPPSGTFLAVGTTTTFAYY548 pKa = 7.87DD549 pKa = 3.36TLDD552 pKa = 3.52NTGGTVTCSYY562 pKa = 11.24DD563 pKa = 3.27VTVVDD568 pKa = 4.3EE569 pKa = 4.75EE570 pKa = 5.19GPTFATQNITLDD582 pKa = 3.42VTMDD586 pKa = 3.46GTVSIVAEE594 pKa = 4.38DD595 pKa = 4.1LLGLDD600 pKa = 5.1LLASDD605 pKa = 4.18YY606 pKa = 11.15TVDD609 pKa = 3.46QTGTFNRR616 pKa = 11.84EE617 pKa = 4.27DD618 pKa = 3.19ISTTGTEE625 pKa = 3.53ISLFDD630 pKa = 4.21DD631 pKa = 3.9QVSSALPIGFEE642 pKa = 3.92FAFYY646 pKa = 10.46GALYY650 pKa = 10.06NDD652 pKa = 5.25FYY654 pKa = 10.94ISSNGFITFSGNSDD668 pKa = 3.83DD669 pKa = 4.73GCCTGQTLPDD679 pKa = 3.3VTEE682 pKa = 4.29PNNLIAFDD690 pKa = 3.81WDD692 pKa = 4.05DD693 pKa = 4.17YY694 pKa = 11.6DD695 pKa = 4.38PEE697 pKa = 5.63SGGTIRR703 pKa = 11.84YY704 pKa = 5.8QTIGTAPNRR713 pKa = 11.84ILIMDD718 pKa = 4.45FDD720 pKa = 4.69AVLHH724 pKa = 6.64IDD726 pKa = 3.77YY727 pKa = 10.94NDD729 pKa = 3.41NTDD732 pKa = 3.04ATTTQVKK739 pKa = 10.15LFEE742 pKa = 4.31GTNRR746 pKa = 11.84IEE748 pKa = 3.75IHH750 pKa = 5.91ATNIPDD756 pKa = 4.23LSNTKK761 pKa = 8.31TQGLEE766 pKa = 4.23NIDD769 pKa = 3.29GTAAIIVPGRR779 pKa = 11.84NAATWSASNDD789 pKa = 3.02VVAFVPVTGGVADD802 pKa = 3.64SCGVDD807 pKa = 3.2TFVASQTTFDD817 pKa = 4.5CSNLGANTVTLTATDD832 pKa = 3.54TNGNITEE839 pKa = 4.77KK840 pKa = 10.49IAIVTVTDD848 pKa = 4.22ANNFCPDD855 pKa = 3.11ILVSPKK861 pKa = 10.36VFLQGAALNPNVGEE875 pKa = 4.12EE876 pKa = 3.94TLMRR880 pKa = 11.84DD881 pKa = 3.98DD882 pKa = 5.64LRR884 pKa = 11.84IAGQIPTTSPYY895 pKa = 10.55TDD897 pKa = 4.25GLTCDD902 pKa = 3.55ASVFTVTGANAIVDD916 pKa = 4.23WVWLEE921 pKa = 3.69LRR923 pKa = 11.84DD924 pKa = 4.79PITSTTIVSSRR935 pKa = 11.84SALLQRR941 pKa = 11.84DD942 pKa = 3.61GDD944 pKa = 3.91IVDD947 pKa = 3.6IDD949 pKa = 4.06GVSTSVIFSEE959 pKa = 5.13VPGAYY964 pKa = 9.97YY965 pKa = 10.65LVVTHH970 pKa = 7.08RR971 pKa = 11.84NHH973 pKa = 6.83LGIRR977 pKa = 11.84SANTYY982 pKa = 9.88NLSSTTTTINLSTSSNSVAGGTNAIIDD1009 pKa = 3.99LGNGKK1014 pKa = 10.09FGLIGGDD1021 pKa = 3.66FDD1023 pKa = 6.99ANGQVQNTDD1032 pKa = 3.04LSGVRR1037 pKa = 11.84PVLGQSQYY1045 pKa = 11.52SAGDD1049 pKa = 3.48LDD1051 pKa = 4.62MNSQVQNTDD1060 pKa = 2.38INNLLNPNTGKK1071 pKa = 10.77GKK1073 pKa = 9.98QFAKK1077 pKa = 10.41QNLRR1081 pKa = 11.84AKK1083 pKa = 10.12LKK1085 pKa = 10.52KK1086 pKa = 10.36
Molecular weight: 114.4 kDa
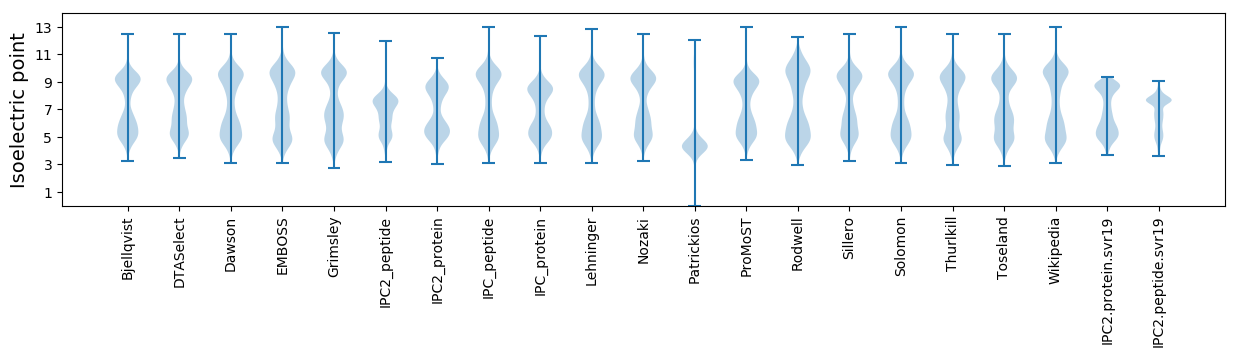
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G5GGQ7|A0A2G5GGQ7_9FLAO Uncharacterized protein OS=Gaetbulibacter sp. 4G1 OX=1891971 GN=BFR04_01035 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1180465 |
50 |
3150 |
353.0 |
39.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.985 ± 0.041 | 0.738 ± 0.014 |
5.673 ± 0.034 | 6.273 ± 0.04 |
5.357 ± 0.038 | 6.269 ± 0.046 |
1.757 ± 0.019 | 8.353 ± 0.041 |
8.259 ± 0.057 | 9.117 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.003 ± 0.02 | 6.895 ± 0.042 |
3.227 ± 0.024 | 3.122 ± 0.022 |
3.173 ± 0.025 | 6.703 ± 0.034 |
5.804 ± 0.05 | 6.024 ± 0.028 |
1.091 ± 0.017 | 4.177 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
