
Rubritalea profundi
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Rubritaleaceae; Rubritalea
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
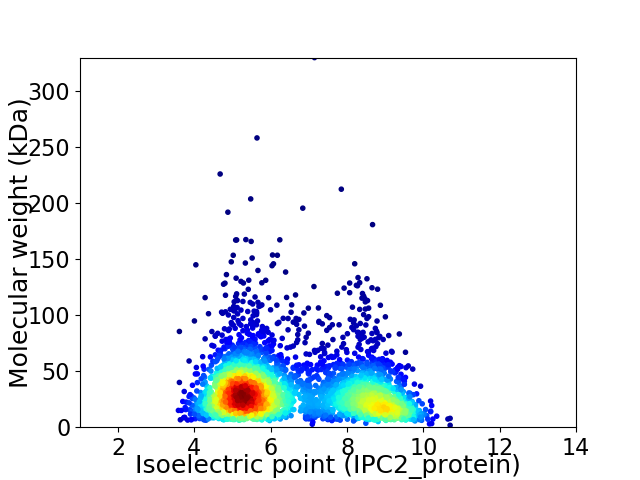
Virtual 2D-PAGE plot for 3389 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7U2U6|A0A2S7U2U6_9BACT Sec-independent protein translocase protein TatC OS=Rubritalea profundi OX=1658618 GN=tatC PE=3 SV=1
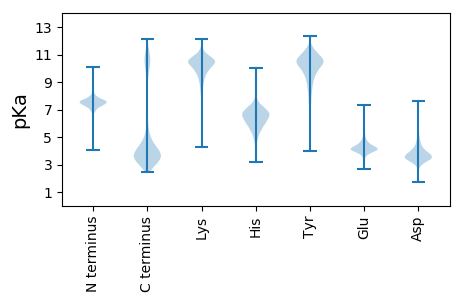
MM1 pKa = 6.84GHH3 pKa = 6.36NMGLSHH9 pKa = 7.61DD10 pKa = 4.13TTTSDD15 pKa = 3.21TIGYY19 pKa = 9.16YY20 pKa = 10.32GGHH23 pKa = 5.71GTGDD27 pKa = 3.9TSWGPIMGTGFNRR40 pKa = 11.84DD41 pKa = 2.8VTQWSKK47 pKa = 10.96GDD49 pKa = 3.56YY50 pKa = 8.21YY51 pKa = 11.63NSYY54 pKa = 10.98NEE56 pKa = 4.22STDD59 pKa = 3.52DD60 pKa = 3.71DD61 pKa = 4.38LAIISARR68 pKa = 11.84VPYY71 pKa = 10.44LADD74 pKa = 5.09DD75 pKa = 4.26YY76 pKa = 11.89GDD78 pKa = 5.57DD79 pKa = 4.5IATATPWFSEE89 pKa = 4.13TDD91 pKa = 3.59GSITQAGLIEE101 pKa = 4.47TTDD104 pKa = 3.75DD105 pKa = 3.82PDD107 pKa = 3.61VFEE110 pKa = 4.91FATGAGTVVFNANTFKK126 pKa = 11.03SDD128 pKa = 3.19AANSHH133 pKa = 5.59TNGGNLDD140 pKa = 3.59INLEE144 pKa = 4.17LRR146 pKa = 11.84DD147 pKa = 3.78SSGTLIALDD156 pKa = 3.93NPALGMNASISQSVAAGNYY175 pKa = 8.31FLFVIPSGAGSPLASSPTGYY195 pKa = 9.18TSYY198 pKa = 11.52GSLGQYY204 pKa = 10.2SITGTIVPTDD214 pKa = 3.74QIALTSPNGGEE225 pKa = 3.28EE226 pKa = 4.17WMRR229 pKa = 11.84STSQPVTWLSGMGGHH244 pKa = 6.67VKK246 pKa = 10.08IEE248 pKa = 4.03LFKK251 pKa = 11.3GGSLDD256 pKa = 3.8SSLAANTPNDD266 pKa = 3.61GLFNWVIPSDD276 pKa = 3.73QAEE279 pKa = 4.27GTDD282 pKa = 3.7YY283 pKa = 10.82KK284 pKa = 10.94IKK286 pKa = 9.37ITSIDD291 pKa = 3.6QPSKK295 pKa = 10.77LDD297 pKa = 3.45KK298 pKa = 11.01SSLNFSVLLEE308 pKa = 4.52SNDD311 pKa = 3.25ILIANMDD318 pKa = 4.39ADD320 pKa = 4.58PGFSTTGLFEE330 pKa = 4.24FGIPSGDD337 pKa = 3.16NDD339 pKa = 4.16AANTNTGTNIYY350 pKa = 9.07DD351 pKa = 3.54TDD353 pKa = 4.39LDD355 pKa = 4.29STCFTASTLTTYY367 pKa = 11.42ALDD370 pKa = 4.13CSNHH374 pKa = 6.17SNVSLEE380 pKa = 4.36FWAHH384 pKa = 5.72TYY386 pKa = 10.37LYY388 pKa = 9.59PDD390 pKa = 3.34YY391 pKa = 9.66TAVFEE396 pKa = 4.64VSNDD400 pKa = 3.59NINWTPLLSQAGMSTRR416 pKa = 11.84SWTKK420 pKa = 8.32YY421 pKa = 9.57SYY423 pKa = 10.82NISSVADD430 pKa = 3.98GQPTVYY436 pKa = 10.31VRR438 pKa = 11.84WSMLGSGTQRR448 pKa = 11.84PGGGLAIDD456 pKa = 4.47DD457 pKa = 4.22AQVVGDD463 pKa = 5.63FVPADD468 pKa = 4.02DD469 pKa = 5.66VSLTSPNGGEE479 pKa = 3.55NWIPNIQNQITWNSSMGGNVKK500 pKa = 9.95IEE502 pKa = 3.95LLEE505 pKa = 4.63GGSVNAVIALSTPNDD520 pKa = 3.59GSYY523 pKa = 11.09LWTAPTAQMLGGDD536 pKa = 3.88YY537 pKa = 10.37KK538 pKa = 11.41VRR540 pKa = 11.84IQSIEE545 pKa = 3.45NTFPYY550 pKa = 10.02RR551 pKa = 11.84RR552 pKa = 11.84EE553 pKa = 3.79
MM1 pKa = 6.84GHH3 pKa = 6.36NMGLSHH9 pKa = 7.61DD10 pKa = 4.13TTTSDD15 pKa = 3.21TIGYY19 pKa = 9.16YY20 pKa = 10.32GGHH23 pKa = 5.71GTGDD27 pKa = 3.9TSWGPIMGTGFNRR40 pKa = 11.84DD41 pKa = 2.8VTQWSKK47 pKa = 10.96GDD49 pKa = 3.56YY50 pKa = 8.21YY51 pKa = 11.63NSYY54 pKa = 10.98NEE56 pKa = 4.22STDD59 pKa = 3.52DD60 pKa = 3.71DD61 pKa = 4.38LAIISARR68 pKa = 11.84VPYY71 pKa = 10.44LADD74 pKa = 5.09DD75 pKa = 4.26YY76 pKa = 11.89GDD78 pKa = 5.57DD79 pKa = 4.5IATATPWFSEE89 pKa = 4.13TDD91 pKa = 3.59GSITQAGLIEE101 pKa = 4.47TTDD104 pKa = 3.75DD105 pKa = 3.82PDD107 pKa = 3.61VFEE110 pKa = 4.91FATGAGTVVFNANTFKK126 pKa = 11.03SDD128 pKa = 3.19AANSHH133 pKa = 5.59TNGGNLDD140 pKa = 3.59INLEE144 pKa = 4.17LRR146 pKa = 11.84DD147 pKa = 3.78SSGTLIALDD156 pKa = 3.93NPALGMNASISQSVAAGNYY175 pKa = 8.31FLFVIPSGAGSPLASSPTGYY195 pKa = 9.18TSYY198 pKa = 11.52GSLGQYY204 pKa = 10.2SITGTIVPTDD214 pKa = 3.74QIALTSPNGGEE225 pKa = 3.28EE226 pKa = 4.17WMRR229 pKa = 11.84STSQPVTWLSGMGGHH244 pKa = 6.67VKK246 pKa = 10.08IEE248 pKa = 4.03LFKK251 pKa = 11.3GGSLDD256 pKa = 3.8SSLAANTPNDD266 pKa = 3.61GLFNWVIPSDD276 pKa = 3.73QAEE279 pKa = 4.27GTDD282 pKa = 3.7YY283 pKa = 10.82KK284 pKa = 10.94IKK286 pKa = 9.37ITSIDD291 pKa = 3.6QPSKK295 pKa = 10.77LDD297 pKa = 3.45KK298 pKa = 11.01SSLNFSVLLEE308 pKa = 4.52SNDD311 pKa = 3.25ILIANMDD318 pKa = 4.39ADD320 pKa = 4.58PGFSTTGLFEE330 pKa = 4.24FGIPSGDD337 pKa = 3.16NDD339 pKa = 4.16AANTNTGTNIYY350 pKa = 9.07DD351 pKa = 3.54TDD353 pKa = 4.39LDD355 pKa = 4.29STCFTASTLTTYY367 pKa = 11.42ALDD370 pKa = 4.13CSNHH374 pKa = 6.17SNVSLEE380 pKa = 4.36FWAHH384 pKa = 5.72TYY386 pKa = 10.37LYY388 pKa = 9.59PDD390 pKa = 3.34YY391 pKa = 9.66TAVFEE396 pKa = 4.64VSNDD400 pKa = 3.59NINWTPLLSQAGMSTRR416 pKa = 11.84SWTKK420 pKa = 8.32YY421 pKa = 9.57SYY423 pKa = 10.82NISSVADD430 pKa = 3.98GQPTVYY436 pKa = 10.31VRR438 pKa = 11.84WSMLGSGTQRR448 pKa = 11.84PGGGLAIDD456 pKa = 4.47DD457 pKa = 4.22AQVVGDD463 pKa = 5.63FVPADD468 pKa = 4.02DD469 pKa = 5.66VSLTSPNGGEE479 pKa = 3.55NWIPNIQNQITWNSSMGGNVKK500 pKa = 9.95IEE502 pKa = 3.95LLEE505 pKa = 4.63GGSVNAVIALSTPNDD520 pKa = 3.59GSYY523 pKa = 11.09LWTAPTAQMLGGDD536 pKa = 3.88YY537 pKa = 10.37KK538 pKa = 11.41VRR540 pKa = 11.84IQSIEE545 pKa = 3.45NTFPYY550 pKa = 10.02RR551 pKa = 11.84RR552 pKa = 11.84EE553 pKa = 3.79
Molecular weight: 59.0 kDa
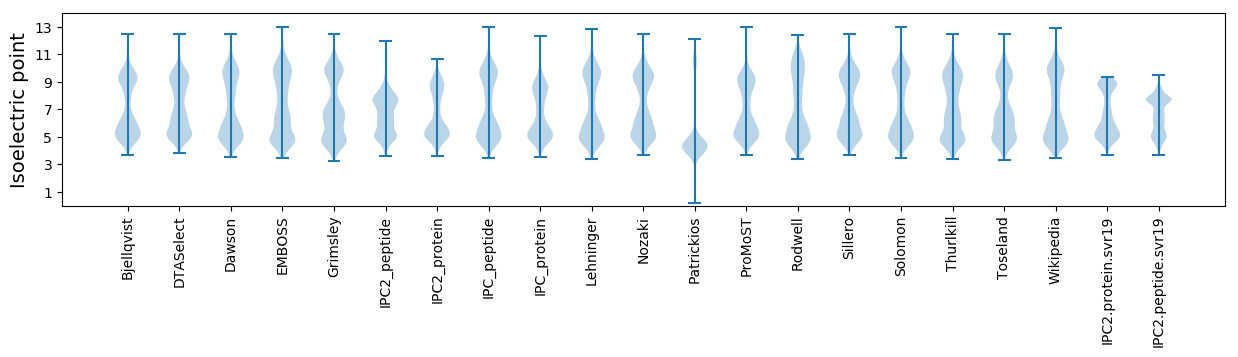
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7U3Q9|A0A2S7U3Q9_9BACT Protein RecA OS=Rubritalea profundi OX=1658618 GN=recA PE=3 SV=1
RR1 pKa = 7.43PCGIGYY7 pKa = 8.04LAPLGSRR14 pKa = 11.84RR15 pKa = 11.84ALSFWQITPAFVSFEE30 pKa = 4.18SLSGIKK36 pKa = 9.91IFFHH40 pKa = 6.33CALRR44 pKa = 11.84GASQRR49 pKa = 11.84INSTKK54 pKa = 9.76PNPYY58 pKa = 9.61SHH60 pKa = 6.42QKK62 pKa = 9.89NLRR65 pKa = 11.84LCGSARR71 pKa = 11.84GFEE74 pKa = 4.53FISRR78 pKa = 11.84AEE80 pKa = 4.08PHH82 pKa = 6.64SKK84 pKa = 8.55TT85 pKa = 3.73
RR1 pKa = 7.43PCGIGYY7 pKa = 8.04LAPLGSRR14 pKa = 11.84RR15 pKa = 11.84ALSFWQITPAFVSFEE30 pKa = 4.18SLSGIKK36 pKa = 9.91IFFHH40 pKa = 6.33CALRR44 pKa = 11.84GASQRR49 pKa = 11.84INSTKK54 pKa = 9.76PNPYY58 pKa = 9.61SHH60 pKa = 6.42QKK62 pKa = 9.89NLRR65 pKa = 11.84LCGSARR71 pKa = 11.84GFEE74 pKa = 4.53FISRR78 pKa = 11.84AEE80 pKa = 4.08PHH82 pKa = 6.64SKK84 pKa = 8.55TT85 pKa = 3.73
Molecular weight: 9.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1083921 |
15 |
2907 |
319.8 |
35.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.847 ± 0.042 | 1.086 ± 0.014 |
5.618 ± 0.032 | 6.291 ± 0.037 |
4.03 ± 0.025 | 7.313 ± 0.041 |
2.299 ± 0.021 | 6.032 ± 0.031 |
6.1 ± 0.045 | 9.899 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.398 ± 0.02 | 3.805 ± 0.029 |
4.443 ± 0.022 | 3.634 ± 0.028 |
4.987 ± 0.032 | 6.859 ± 0.04 |
5.557 ± 0.039 | 6.513 ± 0.038 |
1.415 ± 0.019 | 2.876 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
