
candidate division Zixibacteria bacterium
Taxonomy: cellular organisms; Bacteria; FCB group; candidate division Zixibacteria; unclassified candidate division Zixibacteria
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
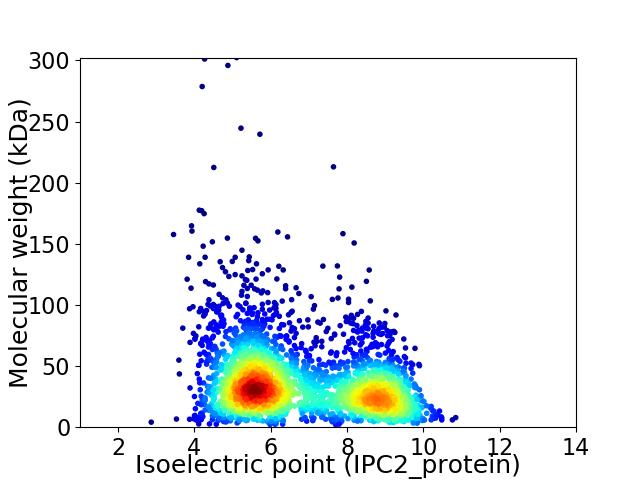
Virtual 2D-PAGE plot for 3031 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I7LZI6|A0A6I7LZI6_9BACT Uncharacterized protein OS=candidate division Zixibacteria bacterium OX=2053527 GN=TRIP_C21254 PE=4 SV=1
MM1 pKa = 7.46FKK3 pKa = 9.81TAFLLTAILSLFGPLKK19 pKa = 10.46VYY21 pKa = 10.7SFEE24 pKa = 5.18PIFNARR30 pKa = 11.84IDD32 pKa = 3.96YY33 pKa = 10.1EE34 pKa = 4.46VGSNPQAICTADD46 pKa = 3.48LNGDD50 pKa = 4.05GKK52 pKa = 11.18ADD54 pKa = 3.7LAVANLEE61 pKa = 4.08SHH63 pKa = 6.93NISILINQGDD73 pKa = 4.12GTFTSAINYY82 pKa = 8.1AAGLYY87 pKa = 7.84PASICTADD95 pKa = 3.41FDD97 pKa = 5.59GNGTADD103 pKa = 3.92LAVTHH108 pKa = 6.8AMDD111 pKa = 4.62SVVILINNGGGSSFMKK127 pKa = 6.85TTYY130 pKa = 10.24YY131 pKa = 10.89AGGVPYY137 pKa = 10.24SIYY140 pKa = 10.92AADD143 pKa = 3.8IDD145 pKa = 4.59GDD147 pKa = 4.07GDD149 pKa = 3.88TDD151 pKa = 4.21LATANYY157 pKa = 9.21VSNSVSILINNGDD170 pKa = 3.92GTFSSPISYY179 pKa = 10.3VVGGDD184 pKa = 2.9AWSICAADD192 pKa = 4.69LDD194 pKa = 4.74ADD196 pKa = 4.05GTTDD200 pKa = 4.2LALAIDD206 pKa = 4.08KK207 pKa = 10.5GVSVLRR213 pKa = 11.84NNGDD217 pKa = 3.71GTFSAPNIYY226 pKa = 10.09EE227 pKa = 3.96VGFTSIFICAVDD239 pKa = 3.84VNATGIIDD247 pKa = 3.97LAVVNHH253 pKa = 6.48NGVTVLKK260 pKa = 10.52NKK262 pKa = 10.36GDD264 pKa = 3.88GTFGALDD271 pKa = 3.74YY272 pKa = 11.24YY273 pKa = 10.37EE274 pKa = 4.86TGSDD278 pKa = 3.26PWSVIAADD286 pKa = 4.02LDD288 pKa = 4.03GDD290 pKa = 4.28GFSDD294 pKa = 4.62LAISHH299 pKa = 5.68IRR301 pKa = 11.84FWGRR305 pKa = 11.84ISILKK310 pKa = 10.31NNGDD314 pKa = 3.78GTFAAAVSFDD324 pKa = 3.98AKK326 pKa = 9.87EE327 pKa = 3.79KK328 pKa = 10.58SYY330 pKa = 11.04SVCAADD336 pKa = 4.32FNGDD340 pKa = 3.51NRR342 pKa = 11.84PDD344 pKa = 3.35LAVANGLWNSISILRR359 pKa = 11.84NGGNAVFPVAPYY371 pKa = 9.78FWVGDD376 pKa = 4.18LPSSAYY382 pKa = 9.63PADD385 pKa = 3.65FDD387 pKa = 4.92GDD389 pKa = 3.99GKK391 pKa = 10.7IDD393 pKa = 4.15LAVLNAGEE401 pKa = 4.05YY402 pKa = 9.76PYY404 pKa = 10.28TEE406 pKa = 3.59RR407 pKa = 11.84VSILKK412 pKa = 10.63NNGNTTFSSADD423 pKa = 3.59SYY425 pKa = 11.87SMEE428 pKa = 4.17HH429 pKa = 6.01FAKK432 pKa = 10.18SVYY435 pKa = 10.55AADD438 pKa = 3.74FDD440 pKa = 5.08GDD442 pKa = 3.77GKK444 pKa = 11.2ADD446 pKa = 3.66LVAANDD452 pKa = 4.31DD453 pKa = 4.01DD454 pKa = 5.19PGTVSLFKK462 pKa = 11.27NNGDD466 pKa = 3.61GTFAAAVNYY475 pKa = 9.64GAGALPYY482 pKa = 9.35NICAADD488 pKa = 3.53FDD490 pKa = 5.3GDD492 pKa = 3.73GSADD496 pKa = 3.33LAVANRR502 pKa = 11.84GNDD505 pKa = 3.12ANYY508 pKa = 10.0PGGFSILKK516 pKa = 10.09NNGDD520 pKa = 3.78GTFAPAVSYY529 pKa = 10.65GAGGGTSICSADD541 pKa = 3.21FDD543 pKa = 5.34GNGKK547 pKa = 10.07ADD549 pKa = 4.0LALSNAGGVSILINNGDD566 pKa = 3.74GTFAAAVNCNTGISPSSIVAVDD588 pKa = 3.79CDD590 pKa = 3.91SDD592 pKa = 3.82GDD594 pKa = 3.92ADD596 pKa = 4.09LAVANRR602 pKa = 11.84GSGSDD607 pKa = 3.61YY608 pKa = 10.51PGNISILINNGDD620 pKa = 3.74GTFAAAVNYY629 pKa = 10.71AEE631 pKa = 4.49GVRR634 pKa = 11.84PSALMVADD642 pKa = 4.69FSGDD646 pKa = 3.61GIADD650 pKa = 3.89LAVKK654 pKa = 10.09NNCDD658 pKa = 3.67VLILICLGDD667 pKa = 3.72GTFVSTLNYY676 pKa = 9.87GVRR679 pKa = 11.84DD680 pKa = 4.05DD681 pKa = 4.39GFGSISAADD690 pKa = 3.95FNNDD694 pKa = 3.28GKK696 pKa = 10.15PDD698 pKa = 3.75LAVTTGGRR706 pKa = 11.84SDD708 pKa = 3.33VQFLFNIAAFVFVCGDD724 pKa = 3.45ADD726 pKa = 3.81RR727 pKa = 11.84SGTAGISDD735 pKa = 3.52ITYY738 pKa = 10.71VIDD741 pKa = 3.59FLYY744 pKa = 10.62RR745 pKa = 11.84FGSAPIPVQAADD757 pKa = 3.62INNSGTVNILDD768 pKa = 3.41ISYY771 pKa = 9.05MINYY775 pKa = 9.34LYY777 pKa = 11.2KK778 pKa = 10.74NGTALNCPP786 pKa = 4.03
MM1 pKa = 7.46FKK3 pKa = 9.81TAFLLTAILSLFGPLKK19 pKa = 10.46VYY21 pKa = 10.7SFEE24 pKa = 5.18PIFNARR30 pKa = 11.84IDD32 pKa = 3.96YY33 pKa = 10.1EE34 pKa = 4.46VGSNPQAICTADD46 pKa = 3.48LNGDD50 pKa = 4.05GKK52 pKa = 11.18ADD54 pKa = 3.7LAVANLEE61 pKa = 4.08SHH63 pKa = 6.93NISILINQGDD73 pKa = 4.12GTFTSAINYY82 pKa = 8.1AAGLYY87 pKa = 7.84PASICTADD95 pKa = 3.41FDD97 pKa = 5.59GNGTADD103 pKa = 3.92LAVTHH108 pKa = 6.8AMDD111 pKa = 4.62SVVILINNGGGSSFMKK127 pKa = 6.85TTYY130 pKa = 10.24YY131 pKa = 10.89AGGVPYY137 pKa = 10.24SIYY140 pKa = 10.92AADD143 pKa = 3.8IDD145 pKa = 4.59GDD147 pKa = 4.07GDD149 pKa = 3.88TDD151 pKa = 4.21LATANYY157 pKa = 9.21VSNSVSILINNGDD170 pKa = 3.92GTFSSPISYY179 pKa = 10.3VVGGDD184 pKa = 2.9AWSICAADD192 pKa = 4.69LDD194 pKa = 4.74ADD196 pKa = 4.05GTTDD200 pKa = 4.2LALAIDD206 pKa = 4.08KK207 pKa = 10.5GVSVLRR213 pKa = 11.84NNGDD217 pKa = 3.71GTFSAPNIYY226 pKa = 10.09EE227 pKa = 3.96VGFTSIFICAVDD239 pKa = 3.84VNATGIIDD247 pKa = 3.97LAVVNHH253 pKa = 6.48NGVTVLKK260 pKa = 10.52NKK262 pKa = 10.36GDD264 pKa = 3.88GTFGALDD271 pKa = 3.74YY272 pKa = 11.24YY273 pKa = 10.37EE274 pKa = 4.86TGSDD278 pKa = 3.26PWSVIAADD286 pKa = 4.02LDD288 pKa = 4.03GDD290 pKa = 4.28GFSDD294 pKa = 4.62LAISHH299 pKa = 5.68IRR301 pKa = 11.84FWGRR305 pKa = 11.84ISILKK310 pKa = 10.31NNGDD314 pKa = 3.78GTFAAAVSFDD324 pKa = 3.98AKK326 pKa = 9.87EE327 pKa = 3.79KK328 pKa = 10.58SYY330 pKa = 11.04SVCAADD336 pKa = 4.32FNGDD340 pKa = 3.51NRR342 pKa = 11.84PDD344 pKa = 3.35LAVANGLWNSISILRR359 pKa = 11.84NGGNAVFPVAPYY371 pKa = 9.78FWVGDD376 pKa = 4.18LPSSAYY382 pKa = 9.63PADD385 pKa = 3.65FDD387 pKa = 4.92GDD389 pKa = 3.99GKK391 pKa = 10.7IDD393 pKa = 4.15LAVLNAGEE401 pKa = 4.05YY402 pKa = 9.76PYY404 pKa = 10.28TEE406 pKa = 3.59RR407 pKa = 11.84VSILKK412 pKa = 10.63NNGNTTFSSADD423 pKa = 3.59SYY425 pKa = 11.87SMEE428 pKa = 4.17HH429 pKa = 6.01FAKK432 pKa = 10.18SVYY435 pKa = 10.55AADD438 pKa = 3.74FDD440 pKa = 5.08GDD442 pKa = 3.77GKK444 pKa = 11.2ADD446 pKa = 3.66LVAANDD452 pKa = 4.31DD453 pKa = 4.01DD454 pKa = 5.19PGTVSLFKK462 pKa = 11.27NNGDD466 pKa = 3.61GTFAAAVNYY475 pKa = 9.64GAGALPYY482 pKa = 9.35NICAADD488 pKa = 3.53FDD490 pKa = 5.3GDD492 pKa = 3.73GSADD496 pKa = 3.33LAVANRR502 pKa = 11.84GNDD505 pKa = 3.12ANYY508 pKa = 10.0PGGFSILKK516 pKa = 10.09NNGDD520 pKa = 3.78GTFAPAVSYY529 pKa = 10.65GAGGGTSICSADD541 pKa = 3.21FDD543 pKa = 5.34GNGKK547 pKa = 10.07ADD549 pKa = 4.0LALSNAGGVSILINNGDD566 pKa = 3.74GTFAAAVNCNTGISPSSIVAVDD588 pKa = 3.79CDD590 pKa = 3.91SDD592 pKa = 3.82GDD594 pKa = 3.92ADD596 pKa = 4.09LAVANRR602 pKa = 11.84GSGSDD607 pKa = 3.61YY608 pKa = 10.51PGNISILINNGDD620 pKa = 3.74GTFAAAVNYY629 pKa = 10.71AEE631 pKa = 4.49GVRR634 pKa = 11.84PSALMVADD642 pKa = 4.69FSGDD646 pKa = 3.61GIADD650 pKa = 3.89LAVKK654 pKa = 10.09NNCDD658 pKa = 3.67VLILICLGDD667 pKa = 3.72GTFVSTLNYY676 pKa = 9.87GVRR679 pKa = 11.84DD680 pKa = 4.05DD681 pKa = 4.39GFGSISAADD690 pKa = 3.95FNNDD694 pKa = 3.28GKK696 pKa = 10.15PDD698 pKa = 3.75LAVTTGGRR706 pKa = 11.84SDD708 pKa = 3.33VQFLFNIAAFVFVCGDD724 pKa = 3.45ADD726 pKa = 3.81RR727 pKa = 11.84SGTAGISDD735 pKa = 3.52ITYY738 pKa = 10.71VIDD741 pKa = 3.59FLYY744 pKa = 10.62RR745 pKa = 11.84FGSAPIPVQAADD757 pKa = 3.62INNSGTVNILDD768 pKa = 3.41ISYY771 pKa = 9.05MINYY775 pKa = 9.34LYY777 pKa = 11.2KK778 pKa = 10.74NGTALNCPP786 pKa = 4.03
Molecular weight: 81.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I7LPB5|A0A6I7LPB5_9BACT Putative Transcriptional regulator TetR family OS=candidate division Zixibacteria bacterium OX=2053527 GN=TRIP_C20142 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 9.0KK12 pKa = 11.08LNDD15 pKa = 3.15HH16 pKa = 6.3GFRR19 pKa = 11.84KK20 pKa = 10.1RR21 pKa = 11.84MATADD26 pKa = 3.39GRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.35RR41 pKa = 11.84LTVSVRR47 pKa = 11.84KK48 pKa = 9.93RR49 pKa = 11.84KK50 pKa = 9.58
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 9.0KK12 pKa = 11.08LNDD15 pKa = 3.15HH16 pKa = 6.3GFRR19 pKa = 11.84KK20 pKa = 10.1RR21 pKa = 11.84MATADD26 pKa = 3.39GRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.35RR41 pKa = 11.84LTVSVRR47 pKa = 11.84KK48 pKa = 9.93RR49 pKa = 11.84KK50 pKa = 9.58
Molecular weight: 5.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1045971 |
20 |
2865 |
345.1 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.147 ± 0.041 | 1.003 ± 0.021 |
5.702 ± 0.037 | 5.79 ± 0.05 |
4.637 ± 0.035 | 7.701 ± 0.042 |
1.8 ± 0.022 | 7.641 ± 0.033 |
5.606 ± 0.046 | 9.574 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.459 ± 0.02 | 4.304 ± 0.034 |
4.409 ± 0.031 | 2.92 ± 0.024 |
5.376 ± 0.043 | 6.467 ± 0.04 |
5.194 ± 0.041 | 6.349 ± 0.039 |
1.134 ± 0.017 | 3.779 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
