
Muricauda ruestringensis (strain DSM 13258 / CIP 107369 / LMG 19739 / B1)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Muricauda; Muricauda ruestringensis
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
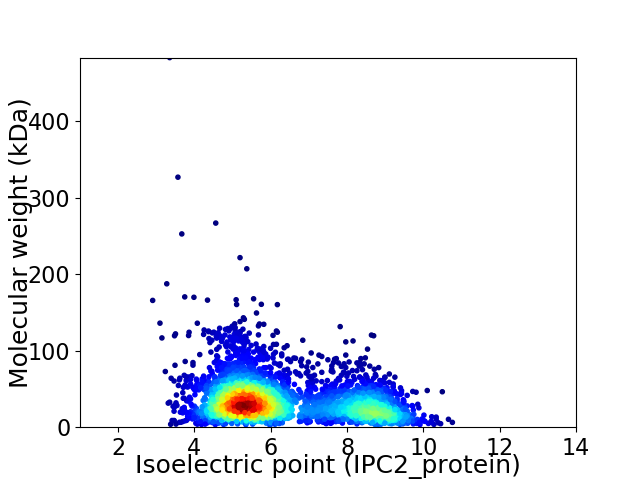
Virtual 2D-PAGE plot for 3425 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
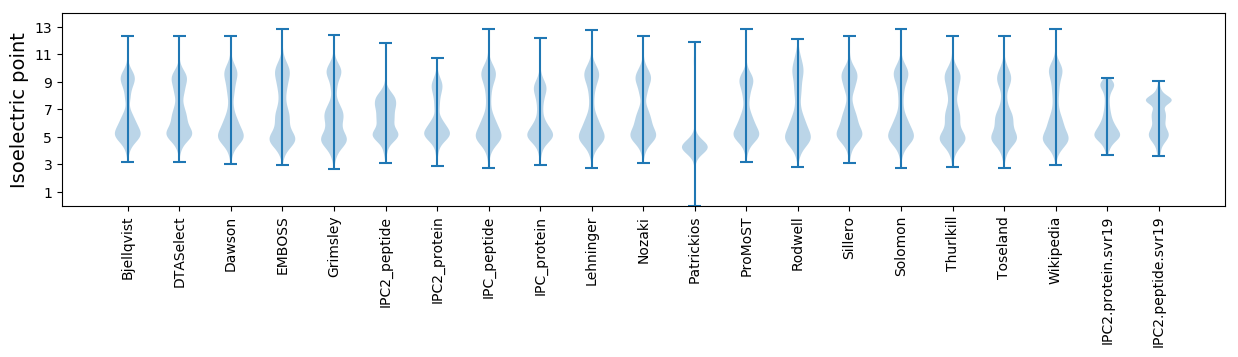
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2PNL4|G2PNL4_MURRD Alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal allergen OS=Muricauda ruestringensis (strain DSM 13258 / CIP 107369 / LMG 19739 / B1) OX=886377 GN=Murru_3415 PE=4 SV=1
MM1 pKa = 7.51LKK3 pKa = 10.86YY4 pKa = 10.51NGNEE8 pKa = 3.99DD9 pKa = 3.32LSGGVTGMMYY19 pKa = 10.74GPDD22 pKa = 3.01NRR24 pKa = 11.84LYY26 pKa = 8.74VTSLKK31 pKa = 10.94GMVNIFTIEE40 pKa = 3.91RR41 pKa = 11.84NLTNDD46 pKa = 3.49YY47 pKa = 10.59EE48 pKa = 4.43VLDD51 pKa = 4.19VEE53 pKa = 4.96VLDD56 pKa = 5.49DD57 pKa = 3.41IQGIVNHH64 pKa = 7.43DD65 pKa = 4.28DD66 pKa = 4.9DD67 pKa = 5.98GSPCNGSYY75 pKa = 9.36SQCHH79 pKa = 4.21TRR81 pKa = 11.84EE82 pKa = 3.92TIGIVLSGSAEE93 pKa = 3.81NPVFYY98 pKa = 10.77VSSSDD103 pKa = 3.41VRR105 pKa = 11.84IGAGSGGGNGDD116 pKa = 3.35VGLDD120 pKa = 3.63TNSGVITRR128 pKa = 11.84FSWTGSSWDD137 pKa = 3.16VVDD140 pKa = 4.54IVRR143 pKa = 11.84GLPRR147 pKa = 11.84SEE149 pKa = 4.39EE150 pKa = 3.77NHH152 pKa = 5.65ATNGLDD158 pKa = 3.61LVTIGGKK165 pKa = 10.02EE166 pKa = 3.96YY167 pKa = 11.23LLVTQGGHH175 pKa = 5.02TNGGGPSVNFVFSTEE190 pKa = 4.02YY191 pKa = 11.21ALAAAILAIDD201 pKa = 4.48LDD203 pKa = 5.34AINSMPVQLDD213 pKa = 2.93NGRR216 pKa = 11.84QYY218 pKa = 10.9IYY220 pKa = 10.67DD221 pKa = 4.85LPTLDD226 pKa = 4.93DD227 pKa = 3.63PTRR230 pKa = 11.84ANVNGITDD238 pKa = 4.47PDD240 pKa = 3.77LPGYY244 pKa = 10.67DD245 pKa = 3.54GVDD248 pKa = 3.5INDD251 pKa = 3.36PWGGNDD257 pKa = 4.09GLNQAKK263 pKa = 9.91VIPNGPVQILSPGYY277 pKa = 9.46RR278 pKa = 11.84NAYY281 pKa = 9.47DD282 pKa = 3.73LVVTKK287 pKa = 10.68SGALYY292 pKa = 9.26VTDD295 pKa = 3.85NGANGGWGGFPVNEE309 pKa = 3.99GTANVNNDD317 pKa = 3.55YY318 pKa = 11.28DD319 pKa = 4.14PNEE322 pKa = 4.92PGSQSYY328 pKa = 10.93SGGEE332 pKa = 3.95HH333 pKa = 6.89INNKK337 pKa = 9.6DD338 pKa = 3.15HH339 pKa = 6.53LQLVTTDD346 pKa = 3.47LSTYY350 pKa = 7.21TFWEE354 pKa = 4.99YY355 pKa = 11.49YY356 pKa = 9.87GGHH359 pKa = 7.1PNPVRR364 pKa = 11.84ANPAGAGLYY373 pKa = 7.41TAPGDD378 pKa = 4.12GNVGAVFRR386 pKa = 11.84TQIFDD391 pKa = 3.75PSSPGAGYY399 pKa = 8.96TSDD402 pKa = 3.88PSIALPVDD410 pKa = 3.98WPPVPVEE417 pKa = 3.74LANVVEE423 pKa = 4.6GDD425 pKa = 2.97WRR427 pKa = 11.84GPDD430 pKa = 3.59EE431 pKa = 5.63DD432 pKa = 5.36NPDD435 pKa = 3.46GPLDD439 pKa = 3.7GEE441 pKa = 4.15IAVWSTNTNGIAEE454 pKa = 4.12YY455 pKa = 10.17RR456 pKa = 11.84ASNFNGAMQGDD467 pKa = 4.93LLATASSGNIRR478 pKa = 11.84RR479 pKa = 11.84VQLTSDD485 pKa = 3.62GQLEE489 pKa = 4.33SLNQSFLSGTKK500 pKa = 10.33GYY502 pKa = 10.66VLAIATTDD510 pKa = 3.57DD511 pKa = 4.67DD512 pKa = 5.26EE513 pKa = 5.04IFPGTIWTGDD523 pKa = 3.43LSGNIQVFEE532 pKa = 4.14PLDD535 pKa = 3.75FVEE538 pKa = 6.34CILPGNPGYY547 pKa = 10.8DD548 pKa = 3.3PLADD552 pKa = 3.87NDD554 pKa = 3.54FDD556 pKa = 5.67GYY558 pKa = 9.65TNQDD562 pKa = 3.78EE563 pKa = 4.72IDD565 pKa = 4.02NGTDD569 pKa = 2.66ICNGGSQPSDD579 pKa = 3.2FDD581 pKa = 3.92SLQGGTLVSDD591 pKa = 4.99LNDD594 pKa = 4.44PDD596 pKa = 5.74DD597 pKa = 5.53DD598 pKa = 4.82FDD600 pKa = 6.15GIPDD604 pKa = 3.48VDD606 pKa = 5.53DD607 pKa = 4.63PFQLGDD613 pKa = 3.54PTTNGSDD620 pKa = 3.35AFTLPVANDD629 pKa = 3.43FFNYY633 pKa = 9.0QQGLGGYY640 pKa = 9.38LGLGLTGFMNNGTGNGNWLNFTDD663 pKa = 5.93RR664 pKa = 11.84RR665 pKa = 11.84DD666 pKa = 3.97DD667 pKa = 4.34PNDD670 pKa = 3.87PNPNDD675 pKa = 3.67VMGGAPGIVTMHH687 pKa = 5.55MTSGTALGTTNTQEE701 pKa = 4.73KK702 pKa = 8.92GLQYY706 pKa = 10.61GAQVDD711 pKa = 3.99VSAGVFRR718 pKa = 11.84VTGGMVGLTGEE729 pKa = 4.12SRR731 pKa = 11.84LYY733 pKa = 11.33GNTAAINGEE742 pKa = 4.13LGFFIGDD749 pKa = 3.93GTQSNYY755 pKa = 10.09IKK757 pKa = 10.63FVVNTNGLLVQQEE770 pKa = 4.46INDD773 pKa = 3.93EE774 pKa = 4.36ANLPITVSIPEE785 pKa = 4.13PNRR788 pKa = 11.84PEE790 pKa = 3.95TGILFHH796 pKa = 6.72FVVNPSTGEE805 pKa = 3.83VTFEE809 pKa = 3.87YY810 pKa = 10.37QIDD813 pKa = 3.75GAAPVEE819 pKa = 4.27IGRR822 pKa = 11.84LTAEE826 pKa = 4.22GSILEE831 pKa = 4.93AIQSQGTDD839 pKa = 3.25LALGLIGTSNTEE851 pKa = 3.98GVEE854 pKa = 5.02LEE856 pKa = 4.4GSWDD860 pKa = 3.55FLNITGSSPTVVQGIADD877 pKa = 3.7MEE879 pKa = 4.71RR880 pKa = 11.84IVGSSDD886 pKa = 2.96EE887 pKa = 4.72TIDD890 pKa = 4.47LDD892 pKa = 4.89TVFDD896 pKa = 4.87DD897 pKa = 4.21NLGVEE902 pKa = 4.25NLSYY906 pKa = 10.9SVEE909 pKa = 4.18NNTNPNIGAIISDD922 pKa = 3.6NMLTITLPATPAEE935 pKa = 4.18STITIRR941 pKa = 11.84ATDD944 pKa = 3.76SEE946 pKa = 4.89SLFVEE951 pKa = 4.12MSFAVSVIEE960 pKa = 5.49DD961 pKa = 3.76NIILYY966 pKa = 9.86RR967 pKa = 11.84INAGGPEE974 pKa = 4.02IASIDD979 pKa = 3.71NDD981 pKa = 4.35LVWSADD987 pKa = 3.35QSSNNSPFLVEE998 pKa = 4.57PGTNTTYY1005 pKa = 11.3AGTITNLDD1013 pKa = 3.54PSIDD1017 pKa = 3.91TNTTPLGIFDD1027 pKa = 3.86TEE1029 pKa = 4.42RR1030 pKa = 11.84FDD1032 pKa = 4.07EE1033 pKa = 5.18ASGAPNMIYY1042 pKa = 10.3SFPVSKK1048 pKa = 10.75NGNYY1052 pKa = 9.41EE1053 pKa = 3.34IHH1055 pKa = 7.56LYY1057 pKa = 9.04MGNGYY1062 pKa = 10.22SGTSQPGEE1070 pKa = 4.16RR1071 pKa = 11.84IFDD1074 pKa = 3.87ALIEE1078 pKa = 5.15GIDD1081 pKa = 4.1LPLLTDD1087 pKa = 3.6IDD1089 pKa = 4.09LSEE1092 pKa = 4.63KK1093 pKa = 10.36FGHH1096 pKa = 6.67ASGGVISHH1104 pKa = 6.64IVKK1107 pKa = 10.34VSDD1110 pKa = 3.48GAIDD1114 pKa = 3.49IEE1116 pKa = 4.41FLHH1119 pKa = 6.88GAIQNPLVNGIEE1131 pKa = 3.98ILDD1134 pKa = 3.93VSDD1137 pKa = 4.22SSTPIYY1143 pKa = 10.95VFDD1146 pKa = 3.56ITNQISSPGEE1156 pKa = 3.73QLNGSLIVDD1165 pKa = 3.97ANGGDD1170 pKa = 3.81GNLTYY1175 pKa = 10.38AAEE1178 pKa = 4.35GLPPGLFIEE1187 pKa = 4.6PTNGQIGGTIEE1198 pKa = 4.12ANAAGGSPYY1207 pKa = 10.43TVIITVDD1214 pKa = 3.58DD1215 pKa = 3.86SDD1217 pKa = 3.94GTTEE1221 pKa = 4.07DD1222 pKa = 3.09TTMVSFQWTIDD1233 pKa = 3.07GDD1235 pKa = 4.43YY1236 pKa = 10.86FWTDD1240 pKa = 2.63KK1241 pKa = 11.58NEE1243 pKa = 4.06NLNYY1247 pKa = 8.21TARR1250 pKa = 11.84HH1251 pKa = 5.6EE1252 pKa = 4.27NSFVQAGDD1260 pKa = 3.01KK1261 pKa = 10.25FYY1263 pKa = 11.45LMGGRR1268 pKa = 11.84EE1269 pKa = 4.01NAQTIDD1275 pKa = 3.12IYY1277 pKa = 11.35DD1278 pKa = 3.88YY1279 pKa = 10.53GTDD1282 pKa = 3.01TWTSLSNSAPFEE1294 pKa = 4.26FNHH1297 pKa = 6.32FQATEE1302 pKa = 3.94YY1303 pKa = 10.72KK1304 pKa = 10.08GLIWIIGSFKK1314 pKa = 10.74TNAFPNEE1321 pKa = 3.77IPAEE1325 pKa = 4.42FIWMFDD1331 pKa = 3.45PASQEE1336 pKa = 4.14WIQGPEE1342 pKa = 3.35IPTNRR1347 pKa = 11.84RR1348 pKa = 11.84RR1349 pKa = 11.84GSSGLVVYY1357 pKa = 9.88QDD1359 pKa = 2.91KK1360 pKa = 10.82FYY1362 pKa = 11.54VVAGNTDD1369 pKa = 3.2GHH1371 pKa = 6.92DD1372 pKa = 3.43GGYY1375 pKa = 10.18VPWFDD1380 pKa = 4.61VYY1382 pKa = 11.43DD1383 pKa = 4.29PSTGTWTALTDD1394 pKa = 3.41APRR1397 pKa = 11.84ARR1399 pKa = 11.84DD1400 pKa = 3.48HH1401 pKa = 6.71FSAVIIGDD1409 pKa = 3.71KK1410 pKa = 10.67LYY1412 pKa = 11.04VAGGRR1417 pKa = 11.84LSGGAGGVWAPTIAEE1432 pKa = 3.86VDD1434 pKa = 3.76VYY1436 pKa = 11.62DD1437 pKa = 4.07FTTGSWSTLPSGQNIPTPRR1456 pKa = 11.84GGAATVNFNNKK1467 pKa = 9.12LVVIGGEE1474 pKa = 4.15VEE1476 pKa = 4.28DD1477 pKa = 4.15EE1478 pKa = 4.34EE1479 pKa = 5.18IYY1481 pKa = 11.11GVLTDD1486 pKa = 4.71DD1487 pKa = 3.85ALKK1490 pKa = 9.26ITEE1493 pKa = 4.76EE1494 pKa = 4.17YY1495 pKa = 10.91DD1496 pKa = 3.51PLSGTWKK1503 pKa = 10.44RR1504 pKa = 11.84LPDD1507 pKa = 3.55MNYY1510 pKa = 9.67EE1511 pKa = 4.04RR1512 pKa = 11.84HH1513 pKa = 5.04GTQAIVSGPGIHH1525 pKa = 6.86ILAGAPNRR1533 pKa = 11.84GGGNQKK1539 pKa = 10.24NMEE1542 pKa = 4.47FLGVDD1547 pKa = 3.57APEE1550 pKa = 4.64GTASQASTLQFPEE1563 pKa = 4.42SVAFEE1568 pKa = 4.66DD1569 pKa = 4.92DD1570 pKa = 3.18QTLEE1574 pKa = 3.89IDD1576 pKa = 4.14LSVMDD1581 pKa = 4.85GNVGMFIRR1589 pKa = 11.84SMEE1592 pKa = 3.84ISGTNASDD1600 pKa = 3.61FSIDD1604 pKa = 3.56SGEE1607 pKa = 4.28LVNALLNPGQTHH1619 pKa = 7.46TITVSLNGAGAGKK1632 pKa = 8.16TATLTIDD1639 pKa = 3.58YY1640 pKa = 9.33GNSSAASIALTGTDD1654 pKa = 3.22VDD1656 pKa = 4.06SEE1658 pKa = 4.85GVVGLTLVDD1667 pKa = 4.11ASTDD1671 pKa = 3.08TDD1673 pKa = 4.16LFNLVDD1679 pKa = 4.07GQQIDD1684 pKa = 3.98LGPSGGQALNIRR1696 pKa = 11.84ANTLGGPGSVLFEE1709 pKa = 4.21LMGPVSATQRR1719 pKa = 11.84EE1720 pKa = 4.8GVAPYY1725 pKa = 11.19ALFGDD1730 pKa = 4.63LSGNYY1735 pKa = 9.57NGRR1738 pKa = 11.84DD1739 pKa = 3.65LPLGNYY1745 pKa = 7.33TLTATAYY1752 pKa = 9.17NGSGSSSGVMGQPLSVNFSLVTGADD1777 pKa = 3.65GTPVAMATADD1787 pKa = 3.82VEE1789 pKa = 4.5TGEE1792 pKa = 5.23APLTVNFTGSNSTDD1806 pKa = 3.24DD1807 pKa = 3.62VGIISYY1813 pKa = 9.75EE1814 pKa = 3.64WDD1816 pKa = 3.73FGDD1819 pKa = 5.68GSALSNEE1826 pKa = 4.23ADD1828 pKa = 3.49PVHH1831 pKa = 7.01TYY1833 pKa = 8.86TVAGSYY1839 pKa = 10.71DD1840 pKa = 3.51AVLTVTDD1847 pKa = 4.73GDD1849 pKa = 4.26GQTDD1853 pKa = 3.59TDD1855 pKa = 4.61TITIIANTVQSDD1867 pKa = 4.09SPISFAEE1874 pKa = 4.02ADD1876 pKa = 4.02QVNGDD1881 pKa = 4.23APLEE1885 pKa = 4.04IQFTGSNSTDD1895 pKa = 3.24DD1896 pKa = 3.62VGIISYY1902 pKa = 9.75EE1903 pKa = 3.64WDD1905 pKa = 3.67FGDD1908 pKa = 5.35GSSLSNEE1915 pKa = 3.94ADD1917 pKa = 3.33PVHH1920 pKa = 7.11TYY1922 pKa = 8.45TAAGSYY1928 pKa = 10.56DD1929 pKa = 3.54AVLTVTDD1936 pKa = 4.73GDD1938 pKa = 4.26GQTDD1942 pKa = 3.54TDD1944 pKa = 4.07FVAINVSGPITEE1956 pKa = 4.62GVVSLTLVDD1965 pKa = 5.07ASADD1969 pKa = 3.33TDD1971 pKa = 4.12LFNLVDD1977 pKa = 4.07GQQIDD1982 pKa = 3.98LGPSGGQALNIRR1994 pKa = 11.84ANTLGGPGSVLFEE2007 pKa = 4.21LMGPVSATQRR2017 pKa = 11.84EE2018 pKa = 4.8GVAPYY2023 pKa = 11.14ALFGDD2028 pKa = 3.6QSGNYY2033 pKa = 8.58NEE2035 pKa = 5.22RR2036 pKa = 11.84DD2037 pKa = 3.68LPLGNYY2043 pKa = 7.33TLTATAYY2050 pKa = 9.17NGSGSSSGIMGQPLTVNFSLVTGTGGLPVAMATADD2085 pKa = 3.82VEE2087 pKa = 4.5TGEE2090 pKa = 5.23APLTVNFTGSDD2101 pKa = 3.43STDD2104 pKa = 3.2DD2105 pKa = 3.52VGIISYY2111 pKa = 9.87GWDD2114 pKa = 3.45FGDD2117 pKa = 5.38GSALSNEE2124 pKa = 4.23ADD2126 pKa = 3.47PVHH2129 pKa = 7.11TYY2131 pKa = 8.45TAAGSYY2137 pKa = 10.93DD2138 pKa = 3.49AMLTVTDD2145 pKa = 4.99GDD2147 pKa = 4.25GQTDD2151 pKa = 3.74TDD2153 pKa = 3.92TVTINVSGPITEE2165 pKa = 4.62GVVSLTLVDD2174 pKa = 4.39ASSDD2178 pKa = 3.22TDD2180 pKa = 4.72LFDD2183 pKa = 5.76LVDD2186 pKa = 4.13GQQIDD2191 pKa = 3.98LGPSGGQSLNIRR2203 pKa = 11.84ANTLGVAGSVLFEE2216 pKa = 3.97LTGPVSATQRR2226 pKa = 11.84EE2227 pKa = 4.8GVAPYY2232 pKa = 11.19ALFGDD2237 pKa = 4.23LSGNYY2242 pKa = 8.73NEE2244 pKa = 5.52RR2245 pKa = 11.84DD2246 pKa = 3.68LPLGNYY2252 pKa = 7.33TLTATAYY2259 pKa = 9.17NGSGSSSGIMGQPLMIDD2276 pKa = 3.67FSIVSGLTGKK2286 pKa = 10.46ASSSNMDD2293 pKa = 3.37NALVEE2298 pKa = 4.55SEE2300 pKa = 4.27PQFKK2304 pKa = 10.81VEE2306 pKa = 4.29SKK2308 pKa = 10.6DD2309 pKa = 3.49VPFEE2313 pKa = 4.83IIMFPNPGATEE2324 pKa = 4.03VTISTNSLNSDD2335 pKa = 3.79LKK2337 pKa = 10.8GVRR2340 pKa = 11.84IFDD2343 pKa = 3.39ATGQLVRR2350 pKa = 11.84SFNPSEE2356 pKa = 3.96YY2357 pKa = 9.82RR2358 pKa = 11.84DD2359 pKa = 3.36GRR2361 pKa = 11.84NDD2363 pKa = 3.03YY2364 pKa = 10.29KK2365 pKa = 11.37LPVNSLQAGVYY2376 pKa = 9.74HH2377 pKa = 6.93LGILTYY2383 pKa = 10.86DD2384 pKa = 3.2GGTYY2388 pKa = 10.27FKK2390 pKa = 10.94QLIIRR2395 pKa = 11.84KK2396 pKa = 8.92
MM1 pKa = 7.51LKK3 pKa = 10.86YY4 pKa = 10.51NGNEE8 pKa = 3.99DD9 pKa = 3.32LSGGVTGMMYY19 pKa = 10.74GPDD22 pKa = 3.01NRR24 pKa = 11.84LYY26 pKa = 8.74VTSLKK31 pKa = 10.94GMVNIFTIEE40 pKa = 3.91RR41 pKa = 11.84NLTNDD46 pKa = 3.49YY47 pKa = 10.59EE48 pKa = 4.43VLDD51 pKa = 4.19VEE53 pKa = 4.96VLDD56 pKa = 5.49DD57 pKa = 3.41IQGIVNHH64 pKa = 7.43DD65 pKa = 4.28DD66 pKa = 4.9DD67 pKa = 5.98GSPCNGSYY75 pKa = 9.36SQCHH79 pKa = 4.21TRR81 pKa = 11.84EE82 pKa = 3.92TIGIVLSGSAEE93 pKa = 3.81NPVFYY98 pKa = 10.77VSSSDD103 pKa = 3.41VRR105 pKa = 11.84IGAGSGGGNGDD116 pKa = 3.35VGLDD120 pKa = 3.63TNSGVITRR128 pKa = 11.84FSWTGSSWDD137 pKa = 3.16VVDD140 pKa = 4.54IVRR143 pKa = 11.84GLPRR147 pKa = 11.84SEE149 pKa = 4.39EE150 pKa = 3.77NHH152 pKa = 5.65ATNGLDD158 pKa = 3.61LVTIGGKK165 pKa = 10.02EE166 pKa = 3.96YY167 pKa = 11.23LLVTQGGHH175 pKa = 5.02TNGGGPSVNFVFSTEE190 pKa = 4.02YY191 pKa = 11.21ALAAAILAIDD201 pKa = 4.48LDD203 pKa = 5.34AINSMPVQLDD213 pKa = 2.93NGRR216 pKa = 11.84QYY218 pKa = 10.9IYY220 pKa = 10.67DD221 pKa = 4.85LPTLDD226 pKa = 4.93DD227 pKa = 3.63PTRR230 pKa = 11.84ANVNGITDD238 pKa = 4.47PDD240 pKa = 3.77LPGYY244 pKa = 10.67DD245 pKa = 3.54GVDD248 pKa = 3.5INDD251 pKa = 3.36PWGGNDD257 pKa = 4.09GLNQAKK263 pKa = 9.91VIPNGPVQILSPGYY277 pKa = 9.46RR278 pKa = 11.84NAYY281 pKa = 9.47DD282 pKa = 3.73LVVTKK287 pKa = 10.68SGALYY292 pKa = 9.26VTDD295 pKa = 3.85NGANGGWGGFPVNEE309 pKa = 3.99GTANVNNDD317 pKa = 3.55YY318 pKa = 11.28DD319 pKa = 4.14PNEE322 pKa = 4.92PGSQSYY328 pKa = 10.93SGGEE332 pKa = 3.95HH333 pKa = 6.89INNKK337 pKa = 9.6DD338 pKa = 3.15HH339 pKa = 6.53LQLVTTDD346 pKa = 3.47LSTYY350 pKa = 7.21TFWEE354 pKa = 4.99YY355 pKa = 11.49YY356 pKa = 9.87GGHH359 pKa = 7.1PNPVRR364 pKa = 11.84ANPAGAGLYY373 pKa = 7.41TAPGDD378 pKa = 4.12GNVGAVFRR386 pKa = 11.84TQIFDD391 pKa = 3.75PSSPGAGYY399 pKa = 8.96TSDD402 pKa = 3.88PSIALPVDD410 pKa = 3.98WPPVPVEE417 pKa = 3.74LANVVEE423 pKa = 4.6GDD425 pKa = 2.97WRR427 pKa = 11.84GPDD430 pKa = 3.59EE431 pKa = 5.63DD432 pKa = 5.36NPDD435 pKa = 3.46GPLDD439 pKa = 3.7GEE441 pKa = 4.15IAVWSTNTNGIAEE454 pKa = 4.12YY455 pKa = 10.17RR456 pKa = 11.84ASNFNGAMQGDD467 pKa = 4.93LLATASSGNIRR478 pKa = 11.84RR479 pKa = 11.84VQLTSDD485 pKa = 3.62GQLEE489 pKa = 4.33SLNQSFLSGTKK500 pKa = 10.33GYY502 pKa = 10.66VLAIATTDD510 pKa = 3.57DD511 pKa = 4.67DD512 pKa = 5.26EE513 pKa = 5.04IFPGTIWTGDD523 pKa = 3.43LSGNIQVFEE532 pKa = 4.14PLDD535 pKa = 3.75FVEE538 pKa = 6.34CILPGNPGYY547 pKa = 10.8DD548 pKa = 3.3PLADD552 pKa = 3.87NDD554 pKa = 3.54FDD556 pKa = 5.67GYY558 pKa = 9.65TNQDD562 pKa = 3.78EE563 pKa = 4.72IDD565 pKa = 4.02NGTDD569 pKa = 2.66ICNGGSQPSDD579 pKa = 3.2FDD581 pKa = 3.92SLQGGTLVSDD591 pKa = 4.99LNDD594 pKa = 4.44PDD596 pKa = 5.74DD597 pKa = 5.53DD598 pKa = 4.82FDD600 pKa = 6.15GIPDD604 pKa = 3.48VDD606 pKa = 5.53DD607 pKa = 4.63PFQLGDD613 pKa = 3.54PTTNGSDD620 pKa = 3.35AFTLPVANDD629 pKa = 3.43FFNYY633 pKa = 9.0QQGLGGYY640 pKa = 9.38LGLGLTGFMNNGTGNGNWLNFTDD663 pKa = 5.93RR664 pKa = 11.84RR665 pKa = 11.84DD666 pKa = 3.97DD667 pKa = 4.34PNDD670 pKa = 3.87PNPNDD675 pKa = 3.67VMGGAPGIVTMHH687 pKa = 5.55MTSGTALGTTNTQEE701 pKa = 4.73KK702 pKa = 8.92GLQYY706 pKa = 10.61GAQVDD711 pKa = 3.99VSAGVFRR718 pKa = 11.84VTGGMVGLTGEE729 pKa = 4.12SRR731 pKa = 11.84LYY733 pKa = 11.33GNTAAINGEE742 pKa = 4.13LGFFIGDD749 pKa = 3.93GTQSNYY755 pKa = 10.09IKK757 pKa = 10.63FVVNTNGLLVQQEE770 pKa = 4.46INDD773 pKa = 3.93EE774 pKa = 4.36ANLPITVSIPEE785 pKa = 4.13PNRR788 pKa = 11.84PEE790 pKa = 3.95TGILFHH796 pKa = 6.72FVVNPSTGEE805 pKa = 3.83VTFEE809 pKa = 3.87YY810 pKa = 10.37QIDD813 pKa = 3.75GAAPVEE819 pKa = 4.27IGRR822 pKa = 11.84LTAEE826 pKa = 4.22GSILEE831 pKa = 4.93AIQSQGTDD839 pKa = 3.25LALGLIGTSNTEE851 pKa = 3.98GVEE854 pKa = 5.02LEE856 pKa = 4.4GSWDD860 pKa = 3.55FLNITGSSPTVVQGIADD877 pKa = 3.7MEE879 pKa = 4.71RR880 pKa = 11.84IVGSSDD886 pKa = 2.96EE887 pKa = 4.72TIDD890 pKa = 4.47LDD892 pKa = 4.89TVFDD896 pKa = 4.87DD897 pKa = 4.21NLGVEE902 pKa = 4.25NLSYY906 pKa = 10.9SVEE909 pKa = 4.18NNTNPNIGAIISDD922 pKa = 3.6NMLTITLPATPAEE935 pKa = 4.18STITIRR941 pKa = 11.84ATDD944 pKa = 3.76SEE946 pKa = 4.89SLFVEE951 pKa = 4.12MSFAVSVIEE960 pKa = 5.49DD961 pKa = 3.76NIILYY966 pKa = 9.86RR967 pKa = 11.84INAGGPEE974 pKa = 4.02IASIDD979 pKa = 3.71NDD981 pKa = 4.35LVWSADD987 pKa = 3.35QSSNNSPFLVEE998 pKa = 4.57PGTNTTYY1005 pKa = 11.3AGTITNLDD1013 pKa = 3.54PSIDD1017 pKa = 3.91TNTTPLGIFDD1027 pKa = 3.86TEE1029 pKa = 4.42RR1030 pKa = 11.84FDD1032 pKa = 4.07EE1033 pKa = 5.18ASGAPNMIYY1042 pKa = 10.3SFPVSKK1048 pKa = 10.75NGNYY1052 pKa = 9.41EE1053 pKa = 3.34IHH1055 pKa = 7.56LYY1057 pKa = 9.04MGNGYY1062 pKa = 10.22SGTSQPGEE1070 pKa = 4.16RR1071 pKa = 11.84IFDD1074 pKa = 3.87ALIEE1078 pKa = 5.15GIDD1081 pKa = 4.1LPLLTDD1087 pKa = 3.6IDD1089 pKa = 4.09LSEE1092 pKa = 4.63KK1093 pKa = 10.36FGHH1096 pKa = 6.67ASGGVISHH1104 pKa = 6.64IVKK1107 pKa = 10.34VSDD1110 pKa = 3.48GAIDD1114 pKa = 3.49IEE1116 pKa = 4.41FLHH1119 pKa = 6.88GAIQNPLVNGIEE1131 pKa = 3.98ILDD1134 pKa = 3.93VSDD1137 pKa = 4.22SSTPIYY1143 pKa = 10.95VFDD1146 pKa = 3.56ITNQISSPGEE1156 pKa = 3.73QLNGSLIVDD1165 pKa = 3.97ANGGDD1170 pKa = 3.81GNLTYY1175 pKa = 10.38AAEE1178 pKa = 4.35GLPPGLFIEE1187 pKa = 4.6PTNGQIGGTIEE1198 pKa = 4.12ANAAGGSPYY1207 pKa = 10.43TVIITVDD1214 pKa = 3.58DD1215 pKa = 3.86SDD1217 pKa = 3.94GTTEE1221 pKa = 4.07DD1222 pKa = 3.09TTMVSFQWTIDD1233 pKa = 3.07GDD1235 pKa = 4.43YY1236 pKa = 10.86FWTDD1240 pKa = 2.63KK1241 pKa = 11.58NEE1243 pKa = 4.06NLNYY1247 pKa = 8.21TARR1250 pKa = 11.84HH1251 pKa = 5.6EE1252 pKa = 4.27NSFVQAGDD1260 pKa = 3.01KK1261 pKa = 10.25FYY1263 pKa = 11.45LMGGRR1268 pKa = 11.84EE1269 pKa = 4.01NAQTIDD1275 pKa = 3.12IYY1277 pKa = 11.35DD1278 pKa = 3.88YY1279 pKa = 10.53GTDD1282 pKa = 3.01TWTSLSNSAPFEE1294 pKa = 4.26FNHH1297 pKa = 6.32FQATEE1302 pKa = 3.94YY1303 pKa = 10.72KK1304 pKa = 10.08GLIWIIGSFKK1314 pKa = 10.74TNAFPNEE1321 pKa = 3.77IPAEE1325 pKa = 4.42FIWMFDD1331 pKa = 3.45PASQEE1336 pKa = 4.14WIQGPEE1342 pKa = 3.35IPTNRR1347 pKa = 11.84RR1348 pKa = 11.84RR1349 pKa = 11.84GSSGLVVYY1357 pKa = 9.88QDD1359 pKa = 2.91KK1360 pKa = 10.82FYY1362 pKa = 11.54VVAGNTDD1369 pKa = 3.2GHH1371 pKa = 6.92DD1372 pKa = 3.43GGYY1375 pKa = 10.18VPWFDD1380 pKa = 4.61VYY1382 pKa = 11.43DD1383 pKa = 4.29PSTGTWTALTDD1394 pKa = 3.41APRR1397 pKa = 11.84ARR1399 pKa = 11.84DD1400 pKa = 3.48HH1401 pKa = 6.71FSAVIIGDD1409 pKa = 3.71KK1410 pKa = 10.67LYY1412 pKa = 11.04VAGGRR1417 pKa = 11.84LSGGAGGVWAPTIAEE1432 pKa = 3.86VDD1434 pKa = 3.76VYY1436 pKa = 11.62DD1437 pKa = 4.07FTTGSWSTLPSGQNIPTPRR1456 pKa = 11.84GGAATVNFNNKK1467 pKa = 9.12LVVIGGEE1474 pKa = 4.15VEE1476 pKa = 4.28DD1477 pKa = 4.15EE1478 pKa = 4.34EE1479 pKa = 5.18IYY1481 pKa = 11.11GVLTDD1486 pKa = 4.71DD1487 pKa = 3.85ALKK1490 pKa = 9.26ITEE1493 pKa = 4.76EE1494 pKa = 4.17YY1495 pKa = 10.91DD1496 pKa = 3.51PLSGTWKK1503 pKa = 10.44RR1504 pKa = 11.84LPDD1507 pKa = 3.55MNYY1510 pKa = 9.67EE1511 pKa = 4.04RR1512 pKa = 11.84HH1513 pKa = 5.04GTQAIVSGPGIHH1525 pKa = 6.86ILAGAPNRR1533 pKa = 11.84GGGNQKK1539 pKa = 10.24NMEE1542 pKa = 4.47FLGVDD1547 pKa = 3.57APEE1550 pKa = 4.64GTASQASTLQFPEE1563 pKa = 4.42SVAFEE1568 pKa = 4.66DD1569 pKa = 4.92DD1570 pKa = 3.18QTLEE1574 pKa = 3.89IDD1576 pKa = 4.14LSVMDD1581 pKa = 4.85GNVGMFIRR1589 pKa = 11.84SMEE1592 pKa = 3.84ISGTNASDD1600 pKa = 3.61FSIDD1604 pKa = 3.56SGEE1607 pKa = 4.28LVNALLNPGQTHH1619 pKa = 7.46TITVSLNGAGAGKK1632 pKa = 8.16TATLTIDD1639 pKa = 3.58YY1640 pKa = 9.33GNSSAASIALTGTDD1654 pKa = 3.22VDD1656 pKa = 4.06SEE1658 pKa = 4.85GVVGLTLVDD1667 pKa = 4.11ASTDD1671 pKa = 3.08TDD1673 pKa = 4.16LFNLVDD1679 pKa = 4.07GQQIDD1684 pKa = 3.98LGPSGGQALNIRR1696 pKa = 11.84ANTLGGPGSVLFEE1709 pKa = 4.21LMGPVSATQRR1719 pKa = 11.84EE1720 pKa = 4.8GVAPYY1725 pKa = 11.19ALFGDD1730 pKa = 4.63LSGNYY1735 pKa = 9.57NGRR1738 pKa = 11.84DD1739 pKa = 3.65LPLGNYY1745 pKa = 7.33TLTATAYY1752 pKa = 9.17NGSGSSSGVMGQPLSVNFSLVTGADD1777 pKa = 3.65GTPVAMATADD1787 pKa = 3.82VEE1789 pKa = 4.5TGEE1792 pKa = 5.23APLTVNFTGSNSTDD1806 pKa = 3.24DD1807 pKa = 3.62VGIISYY1813 pKa = 9.75EE1814 pKa = 3.64WDD1816 pKa = 3.73FGDD1819 pKa = 5.68GSALSNEE1826 pKa = 4.23ADD1828 pKa = 3.49PVHH1831 pKa = 7.01TYY1833 pKa = 8.86TVAGSYY1839 pKa = 10.71DD1840 pKa = 3.51AVLTVTDD1847 pKa = 4.73GDD1849 pKa = 4.26GQTDD1853 pKa = 3.59TDD1855 pKa = 4.61TITIIANTVQSDD1867 pKa = 4.09SPISFAEE1874 pKa = 4.02ADD1876 pKa = 4.02QVNGDD1881 pKa = 4.23APLEE1885 pKa = 4.04IQFTGSNSTDD1895 pKa = 3.24DD1896 pKa = 3.62VGIISYY1902 pKa = 9.75EE1903 pKa = 3.64WDD1905 pKa = 3.67FGDD1908 pKa = 5.35GSSLSNEE1915 pKa = 3.94ADD1917 pKa = 3.33PVHH1920 pKa = 7.11TYY1922 pKa = 8.45TAAGSYY1928 pKa = 10.56DD1929 pKa = 3.54AVLTVTDD1936 pKa = 4.73GDD1938 pKa = 4.26GQTDD1942 pKa = 3.54TDD1944 pKa = 4.07FVAINVSGPITEE1956 pKa = 4.62GVVSLTLVDD1965 pKa = 5.07ASADD1969 pKa = 3.33TDD1971 pKa = 4.12LFNLVDD1977 pKa = 4.07GQQIDD1982 pKa = 3.98LGPSGGQALNIRR1994 pKa = 11.84ANTLGGPGSVLFEE2007 pKa = 4.21LMGPVSATQRR2017 pKa = 11.84EE2018 pKa = 4.8GVAPYY2023 pKa = 11.14ALFGDD2028 pKa = 3.6QSGNYY2033 pKa = 8.58NEE2035 pKa = 5.22RR2036 pKa = 11.84DD2037 pKa = 3.68LPLGNYY2043 pKa = 7.33TLTATAYY2050 pKa = 9.17NGSGSSSGIMGQPLTVNFSLVTGTGGLPVAMATADD2085 pKa = 3.82VEE2087 pKa = 4.5TGEE2090 pKa = 5.23APLTVNFTGSDD2101 pKa = 3.43STDD2104 pKa = 3.2DD2105 pKa = 3.52VGIISYY2111 pKa = 9.87GWDD2114 pKa = 3.45FGDD2117 pKa = 5.38GSALSNEE2124 pKa = 4.23ADD2126 pKa = 3.47PVHH2129 pKa = 7.11TYY2131 pKa = 8.45TAAGSYY2137 pKa = 10.93DD2138 pKa = 3.49AMLTVTDD2145 pKa = 4.99GDD2147 pKa = 4.25GQTDD2151 pKa = 3.74TDD2153 pKa = 3.92TVTINVSGPITEE2165 pKa = 4.62GVVSLTLVDD2174 pKa = 4.39ASSDD2178 pKa = 3.22TDD2180 pKa = 4.72LFDD2183 pKa = 5.76LVDD2186 pKa = 4.13GQQIDD2191 pKa = 3.98LGPSGGQSLNIRR2203 pKa = 11.84ANTLGVAGSVLFEE2216 pKa = 3.97LTGPVSATQRR2226 pKa = 11.84EE2227 pKa = 4.8GVAPYY2232 pKa = 11.19ALFGDD2237 pKa = 4.23LSGNYY2242 pKa = 8.73NEE2244 pKa = 5.52RR2245 pKa = 11.84DD2246 pKa = 3.68LPLGNYY2252 pKa = 7.33TLTATAYY2259 pKa = 9.17NGSGSSSGIMGQPLMIDD2276 pKa = 3.67FSIVSGLTGKK2286 pKa = 10.46ASSSNMDD2293 pKa = 3.37NALVEE2298 pKa = 4.55SEE2300 pKa = 4.27PQFKK2304 pKa = 10.81VEE2306 pKa = 4.29SKK2308 pKa = 10.6DD2309 pKa = 3.49VPFEE2313 pKa = 4.83IIMFPNPGATEE2324 pKa = 4.03VTISTNSLNSDD2335 pKa = 3.79LKK2337 pKa = 10.8GVRR2340 pKa = 11.84IFDD2343 pKa = 3.39ATGQLVRR2350 pKa = 11.84SFNPSEE2356 pKa = 3.96YY2357 pKa = 9.82RR2358 pKa = 11.84DD2359 pKa = 3.36GRR2361 pKa = 11.84NDD2363 pKa = 3.03YY2364 pKa = 10.29KK2365 pKa = 11.37LPVNSLQAGVYY2376 pKa = 9.74HH2377 pKa = 6.93LGILTYY2383 pKa = 10.86DD2384 pKa = 3.2GGTYY2388 pKa = 10.27FKK2390 pKa = 10.94QLIIRR2395 pKa = 11.84KK2396 pKa = 8.92
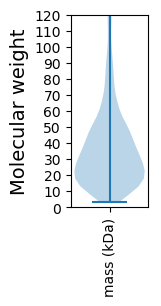
Molecular weight: 252.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2PS07|G2PS07_MURRD SSS sodium solute transporter superfamily OS=Muricauda ruestringensis (strain DSM 13258 / CIP 107369 / LMG 19739 / B1) OX=886377 GN=Murru_0546 PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.36KK31 pKa = 10.55SLLAPIKK38 pKa = 9.77KK39 pKa = 8.55TGGRR43 pKa = 11.84NSQGKK48 pKa = 6.1MTMRR52 pKa = 11.84HH53 pKa = 5.75KK54 pKa = 11.07GGGHH58 pKa = 5.24KK59 pKa = 9.72RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.84FKK68 pKa = 11.0RR69 pKa = 11.84DD70 pKa = 3.56KK71 pKa = 10.86QDD73 pKa = 2.38VDD75 pKa = 3.46ATVVSIQYY83 pKa = 10.16DD84 pKa = 3.6PNRR87 pKa = 11.84TAFIALVEE95 pKa = 4.31YY96 pKa = 10.85KK97 pKa = 10.38DD98 pKa = 3.54GEE100 pKa = 4.12KK101 pKa = 10.26RR102 pKa = 11.84YY103 pKa = 10.29VVAQNGLQVGQTIKK117 pKa = 10.55SGSGSAPEE125 pKa = 4.55IGNALPLGEE134 pKa = 4.94IPLGTIVSCMEE145 pKa = 3.89LRR147 pKa = 11.84PGQGAIMARR156 pKa = 11.84SAGTFAQLMAKK167 pKa = 10.03DD168 pKa = 4.13GKK170 pKa = 10.62FVTLKK175 pKa = 10.58LPSGEE180 pKa = 3.94TRR182 pKa = 11.84MILATCLATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.25GYY255 pKa = 7.08RR256 pKa = 11.84TRR258 pKa = 11.84SRR260 pKa = 11.84TKK262 pKa = 9.51DD263 pKa = 3.07TNKK266 pKa = 10.63YY267 pKa = 9.38IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.36KK31 pKa = 10.55SLLAPIKK38 pKa = 9.77KK39 pKa = 8.55TGGRR43 pKa = 11.84NSQGKK48 pKa = 6.1MTMRR52 pKa = 11.84HH53 pKa = 5.75KK54 pKa = 11.07GGGHH58 pKa = 5.24KK59 pKa = 9.72RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.84FKK68 pKa = 11.0RR69 pKa = 11.84DD70 pKa = 3.56KK71 pKa = 10.86QDD73 pKa = 2.38VDD75 pKa = 3.46ATVVSIQYY83 pKa = 10.16DD84 pKa = 3.6PNRR87 pKa = 11.84TAFIALVEE95 pKa = 4.31YY96 pKa = 10.85KK97 pKa = 10.38DD98 pKa = 3.54GEE100 pKa = 4.12KK101 pKa = 10.26RR102 pKa = 11.84YY103 pKa = 10.29VVAQNGLQVGQTIKK117 pKa = 10.55SGSGSAPEE125 pKa = 4.55IGNALPLGEE134 pKa = 4.94IPLGTIVSCMEE145 pKa = 3.89LRR147 pKa = 11.84PGQGAIMARR156 pKa = 11.84SAGTFAQLMAKK167 pKa = 10.03DD168 pKa = 4.13GKK170 pKa = 10.62FVTLKK175 pKa = 10.58LPSGEE180 pKa = 3.94TRR182 pKa = 11.84MILATCLATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.25GYY255 pKa = 7.08RR256 pKa = 11.84TRR258 pKa = 11.84SRR260 pKa = 11.84TKK262 pKa = 9.51DD263 pKa = 3.07TNKK266 pKa = 10.63YY267 pKa = 9.38IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1152300 |
30 |
4538 |
336.4 |
37.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.45 ± 0.034 | 0.726 ± 0.012 |
5.88 ± 0.032 | 6.955 ± 0.042 |
5.078 ± 0.032 | 6.93 ± 0.038 |
1.853 ± 0.021 | 7.151 ± 0.039 |
7.302 ± 0.054 | 9.396 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.451 ± 0.022 | 5.53 ± 0.035 |
3.629 ± 0.023 | 3.475 ± 0.023 |
3.691 ± 0.026 | 6.316 ± 0.033 |
5.607 ± 0.048 | 6.5 ± 0.03 |
1.125 ± 0.018 | 3.956 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
