
Sphingomonas sp. HDW15A
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
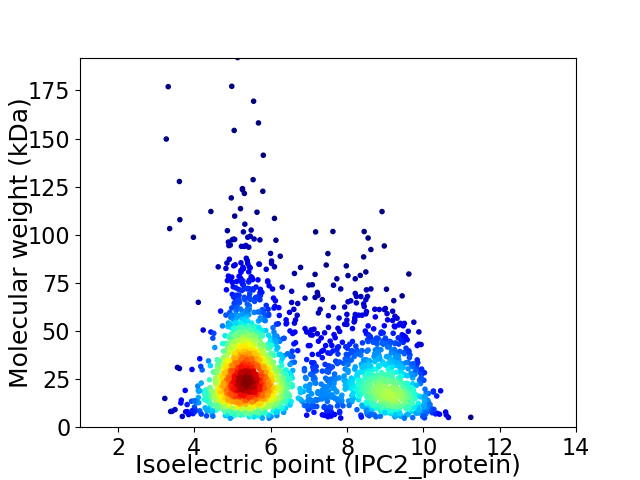
Virtual 2D-PAGE plot for 2226 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G7ZJK4|A0A6G7ZJK4_9SPHN DNA polymerase Y family protein OS=Sphingomonas sp. HDW15A OX=2714942 GN=G7076_00005 PE=3 SV=1
MM1 pKa = 7.38RR2 pKa = 11.84TFSRR6 pKa = 11.84RR7 pKa = 11.84NRR9 pKa = 11.84AACSILAMATVFTVGATPAAAQVAGTGNFSTGTGTINPGAGTVDD53 pKa = 3.03ITGGGQAVIDD63 pKa = 3.89WNATGTVVGGVNQFLAPGNTLTFSSSSDD91 pKa = 3.25FAVLNNVSTGTDD103 pKa = 3.15MLGLNGTVTSSVDD116 pKa = 3.28GVQGRR121 pKa = 11.84GSVYY125 pKa = 9.91FYY127 pKa = 10.59NPNGIVVGGSSVFNVGSLVLSTSPILVSGGAFIDD161 pKa = 4.3GNNQVVFQQGLAGASIDD178 pKa = 3.94VQSGAQINATSNNSYY193 pKa = 10.28VALVAPRR200 pKa = 11.84VVHH203 pKa = 6.63NGTINVNGSAALVGAEE219 pKa = 4.1AANITFSPDD228 pKa = 2.72GLFDD232 pKa = 3.75IEE234 pKa = 4.43VTAGTTDD241 pKa = 3.13ANGVVIGGTITGPEE255 pKa = 3.93AGVGEE260 pKa = 4.41TQRR263 pKa = 11.84IYY265 pKa = 11.39AVAVPKK271 pKa = 10.06NQAMTLAITAGAQLGFDD288 pKa = 2.97IAGAVNVDD296 pKa = 3.05GNAIVLSAGRR306 pKa = 11.84NVTNGDD312 pKa = 3.29ADD314 pKa = 4.1TARR317 pKa = 11.84SAAMGSGTASINSNDD332 pKa = 3.62LTTTSAITASAVNNNIFLGFNDD354 pKa = 3.16ITMASDD360 pKa = 3.25VTIIGEE366 pKa = 4.26DD367 pKa = 3.55MVHH370 pKa = 6.29LRR372 pKa = 11.84AIGANADD379 pKa = 3.77LTVGGLLTMSGDD391 pKa = 3.7VFVAAGQAGQAGDD404 pKa = 3.57VMVSAEE410 pKa = 4.12GGALLTASGGAVLSAVARR428 pKa = 11.84GQASGTAGVNGGNAQGGDD446 pKa = 3.51INVLGSDD453 pKa = 3.74GTLVIAGDD461 pKa = 4.05LSVDD465 pKa = 3.07ANAYY469 pKa = 10.06GGEE472 pKa = 4.14TLVNGANAGSAVGGLIAITAYY493 pKa = 10.5GPDD496 pKa = 3.47GTLSINGGLFATADD510 pKa = 3.72GVGGNGGYY518 pKa = 9.47SEE520 pKa = 5.43SCFSCNGAGGNGQGGQIYY538 pKa = 8.89FQTVGNGGFLTIGDD552 pKa = 5.31DD553 pKa = 3.68IQASASGSGGSSRR566 pKa = 11.84TATGRR571 pKa = 11.84GDD573 pKa = 3.4GGTISATYY581 pKa = 8.99GANGTIQFDD590 pKa = 3.91ANASFTATGAGGQSDD605 pKa = 4.13ALGVDD610 pKa = 3.78GGYY613 pKa = 11.21GKK615 pKa = 11.06GGTVKK620 pKa = 10.72FGAQNNATGGSLTVAGLAYY639 pKa = 10.2AAIEE643 pKa = 4.78GYY645 pKa = 10.88GGDD648 pKa = 3.61SYY650 pKa = 12.04GAGTGNGGTGEE661 pKa = 4.84GGLSDD666 pKa = 3.46NGGNAGTINFQGGLQILGRR685 pKa = 11.84GFGGDD690 pKa = 3.65GNNGTGGQGLGGQGWIDD707 pKa = 3.27AFGANVTIGGDD718 pKa = 3.31YY719 pKa = 10.5DD720 pKa = 4.21INVSGLGGSGITGGFGRR737 pKa = 11.84GGNAYY742 pKa = 9.48LASSGTFLLTGNADD756 pKa = 3.56LVADD760 pKa = 4.25GRR762 pKa = 11.84GGGAFGGNGGLGQGGLANVRR782 pKa = 11.84ATGSGTLTVQGNVDD796 pKa = 3.23AHH798 pKa = 5.97ATGQGGFVAFGSGTAGDD815 pKa = 3.95GDD817 pKa = 3.94GGVVSIHH824 pKa = 6.71AGNGGATPSGGTLNILGSLTGVSDD848 pKa = 5.01GIGGLADD855 pKa = 3.48TATSVGGTGRR865 pKa = 11.84NAEE868 pKa = 4.45LFLFAAGGGDD878 pKa = 3.66LSVGGSTDD886 pKa = 3.45LSADD890 pKa = 3.47GLAGFYY896 pKa = 10.45LEE898 pKa = 5.74CLDD901 pKa = 4.96CGGTPGIGDD910 pKa = 4.03GGNLNISADD919 pKa = 3.9GAGSTLSFGDD929 pKa = 4.67DD930 pKa = 3.34LTATADD936 pKa = 3.52GRR938 pKa = 11.84GGLSSTDD945 pKa = 3.41GGTGGYY951 pKa = 10.58GEE953 pKa = 5.01GGNAGMFAGNGGTITATYY971 pKa = 10.52VDD973 pKa = 4.12ISAIGTGGFDD983 pKa = 3.71YY984 pKa = 11.33NGVGGGAGKK993 pKa = 10.55GGNVYY998 pKa = 10.43FNTNATGVNTITVTGQVFLDD1018 pKa = 3.39ASGYY1022 pKa = 10.8GGGSYY1027 pKa = 10.36GGNGGAGTGGVAGMFVQTGTVTLGSADD1054 pKa = 3.0ISATGEE1060 pKa = 3.98GGTVIEE1066 pKa = 4.55GTGTGGDD1073 pKa = 3.49ATGGSARR1080 pKa = 11.84LQGAGGDD1087 pKa = 3.48ILIAGNLAMDD1097 pKa = 4.53GSAIGGNGTAGGNAIALADD1116 pKa = 3.71LTVNEE1121 pKa = 4.62TNPIAVGVYY1130 pKa = 10.01ANNNGSVTVNGAVAIDD1146 pKa = 3.99ASATGGNGSAGNGGNATAGEE1166 pKa = 4.06IDD1168 pKa = 3.13IVAFFGDD1175 pKa = 3.43IAMGDD1180 pKa = 3.83LAVDD1184 pKa = 3.63ISGNGGDD1191 pKa = 4.52GGQGGNGGNATGGYY1205 pKa = 10.53ADD1207 pKa = 3.52IAFGLGAAAIGGTLTLASATVVADD1231 pKa = 4.67GIGGDD1236 pKa = 3.87GGAGVDD1242 pKa = 4.42GNTGGNGGNGGTGTGGRR1259 pKa = 11.84ISFVGSAAGGFLTSGITVLSAGATGGDD1286 pKa = 3.52GGVGGNGDD1294 pKa = 3.84TGTGGNGGTGGAAFSGSIQSGTVSASGAPGSGGGFDD1330 pKa = 3.64VTSLFAATNAAGGGAGRR1347 pKa = 11.84PGLAAAATEE1356 pKa = 4.39TAATAAVRR1364 pKa = 11.84KK1365 pKa = 9.71VDD1367 pKa = 3.78RR1368 pKa = 11.84QASS1371 pKa = 3.49
MM1 pKa = 7.38RR2 pKa = 11.84TFSRR6 pKa = 11.84RR7 pKa = 11.84NRR9 pKa = 11.84AACSILAMATVFTVGATPAAAQVAGTGNFSTGTGTINPGAGTVDD53 pKa = 3.03ITGGGQAVIDD63 pKa = 3.89WNATGTVVGGVNQFLAPGNTLTFSSSSDD91 pKa = 3.25FAVLNNVSTGTDD103 pKa = 3.15MLGLNGTVTSSVDD116 pKa = 3.28GVQGRR121 pKa = 11.84GSVYY125 pKa = 9.91FYY127 pKa = 10.59NPNGIVVGGSSVFNVGSLVLSTSPILVSGGAFIDD161 pKa = 4.3GNNQVVFQQGLAGASIDD178 pKa = 3.94VQSGAQINATSNNSYY193 pKa = 10.28VALVAPRR200 pKa = 11.84VVHH203 pKa = 6.63NGTINVNGSAALVGAEE219 pKa = 4.1AANITFSPDD228 pKa = 2.72GLFDD232 pKa = 3.75IEE234 pKa = 4.43VTAGTTDD241 pKa = 3.13ANGVVIGGTITGPEE255 pKa = 3.93AGVGEE260 pKa = 4.41TQRR263 pKa = 11.84IYY265 pKa = 11.39AVAVPKK271 pKa = 10.06NQAMTLAITAGAQLGFDD288 pKa = 2.97IAGAVNVDD296 pKa = 3.05GNAIVLSAGRR306 pKa = 11.84NVTNGDD312 pKa = 3.29ADD314 pKa = 4.1TARR317 pKa = 11.84SAAMGSGTASINSNDD332 pKa = 3.62LTTTSAITASAVNNNIFLGFNDD354 pKa = 3.16ITMASDD360 pKa = 3.25VTIIGEE366 pKa = 4.26DD367 pKa = 3.55MVHH370 pKa = 6.29LRR372 pKa = 11.84AIGANADD379 pKa = 3.77LTVGGLLTMSGDD391 pKa = 3.7VFVAAGQAGQAGDD404 pKa = 3.57VMVSAEE410 pKa = 4.12GGALLTASGGAVLSAVARR428 pKa = 11.84GQASGTAGVNGGNAQGGDD446 pKa = 3.51INVLGSDD453 pKa = 3.74GTLVIAGDD461 pKa = 4.05LSVDD465 pKa = 3.07ANAYY469 pKa = 10.06GGEE472 pKa = 4.14TLVNGANAGSAVGGLIAITAYY493 pKa = 10.5GPDD496 pKa = 3.47GTLSINGGLFATADD510 pKa = 3.72GVGGNGGYY518 pKa = 9.47SEE520 pKa = 5.43SCFSCNGAGGNGQGGQIYY538 pKa = 8.89FQTVGNGGFLTIGDD552 pKa = 5.31DD553 pKa = 3.68IQASASGSGGSSRR566 pKa = 11.84TATGRR571 pKa = 11.84GDD573 pKa = 3.4GGTISATYY581 pKa = 8.99GANGTIQFDD590 pKa = 3.91ANASFTATGAGGQSDD605 pKa = 4.13ALGVDD610 pKa = 3.78GGYY613 pKa = 11.21GKK615 pKa = 11.06GGTVKK620 pKa = 10.72FGAQNNATGGSLTVAGLAYY639 pKa = 10.2AAIEE643 pKa = 4.78GYY645 pKa = 10.88GGDD648 pKa = 3.61SYY650 pKa = 12.04GAGTGNGGTGEE661 pKa = 4.84GGLSDD666 pKa = 3.46NGGNAGTINFQGGLQILGRR685 pKa = 11.84GFGGDD690 pKa = 3.65GNNGTGGQGLGGQGWIDD707 pKa = 3.27AFGANVTIGGDD718 pKa = 3.31YY719 pKa = 10.5DD720 pKa = 4.21INVSGLGGSGITGGFGRR737 pKa = 11.84GGNAYY742 pKa = 9.48LASSGTFLLTGNADD756 pKa = 3.56LVADD760 pKa = 4.25GRR762 pKa = 11.84GGGAFGGNGGLGQGGLANVRR782 pKa = 11.84ATGSGTLTVQGNVDD796 pKa = 3.23AHH798 pKa = 5.97ATGQGGFVAFGSGTAGDD815 pKa = 3.95GDD817 pKa = 3.94GGVVSIHH824 pKa = 6.71AGNGGATPSGGTLNILGSLTGVSDD848 pKa = 5.01GIGGLADD855 pKa = 3.48TATSVGGTGRR865 pKa = 11.84NAEE868 pKa = 4.45LFLFAAGGGDD878 pKa = 3.66LSVGGSTDD886 pKa = 3.45LSADD890 pKa = 3.47GLAGFYY896 pKa = 10.45LEE898 pKa = 5.74CLDD901 pKa = 4.96CGGTPGIGDD910 pKa = 4.03GGNLNISADD919 pKa = 3.9GAGSTLSFGDD929 pKa = 4.67DD930 pKa = 3.34LTATADD936 pKa = 3.52GRR938 pKa = 11.84GGLSSTDD945 pKa = 3.41GGTGGYY951 pKa = 10.58GEE953 pKa = 5.01GGNAGMFAGNGGTITATYY971 pKa = 10.52VDD973 pKa = 4.12ISAIGTGGFDD983 pKa = 3.71YY984 pKa = 11.33NGVGGGAGKK993 pKa = 10.55GGNVYY998 pKa = 10.43FNTNATGVNTITVTGQVFLDD1018 pKa = 3.39ASGYY1022 pKa = 10.8GGGSYY1027 pKa = 10.36GGNGGAGTGGVAGMFVQTGTVTLGSADD1054 pKa = 3.0ISATGEE1060 pKa = 3.98GGTVIEE1066 pKa = 4.55GTGTGGDD1073 pKa = 3.49ATGGSARR1080 pKa = 11.84LQGAGGDD1087 pKa = 3.48ILIAGNLAMDD1097 pKa = 4.53GSAIGGNGTAGGNAIALADD1116 pKa = 3.71LTVNEE1121 pKa = 4.62TNPIAVGVYY1130 pKa = 10.01ANNNGSVTVNGAVAIDD1146 pKa = 3.99ASATGGNGSAGNGGNATAGEE1166 pKa = 4.06IDD1168 pKa = 3.13IVAFFGDD1175 pKa = 3.43IAMGDD1180 pKa = 3.83LAVDD1184 pKa = 3.63ISGNGGDD1191 pKa = 4.52GGQGGNGGNATGGYY1205 pKa = 10.53ADD1207 pKa = 3.52IAFGLGAAAIGGTLTLASATVVADD1231 pKa = 4.67GIGGDD1236 pKa = 3.87GGAGVDD1242 pKa = 4.42GNTGGNGGNGGTGTGGRR1259 pKa = 11.84ISFVGSAAGGFLTSGITVLSAGATGGDD1286 pKa = 3.52GGVGGNGDD1294 pKa = 3.84TGTGGNGGTGGAAFSGSIQSGTVSASGAPGSGGGFDD1330 pKa = 3.64VTSLFAATNAAGGGAGRR1347 pKa = 11.84PGLAAAATEE1356 pKa = 4.39TAATAAVRR1364 pKa = 11.84KK1365 pKa = 9.71VDD1367 pKa = 3.78RR1368 pKa = 11.84QASS1371 pKa = 3.49
Molecular weight: 127.71 kDa
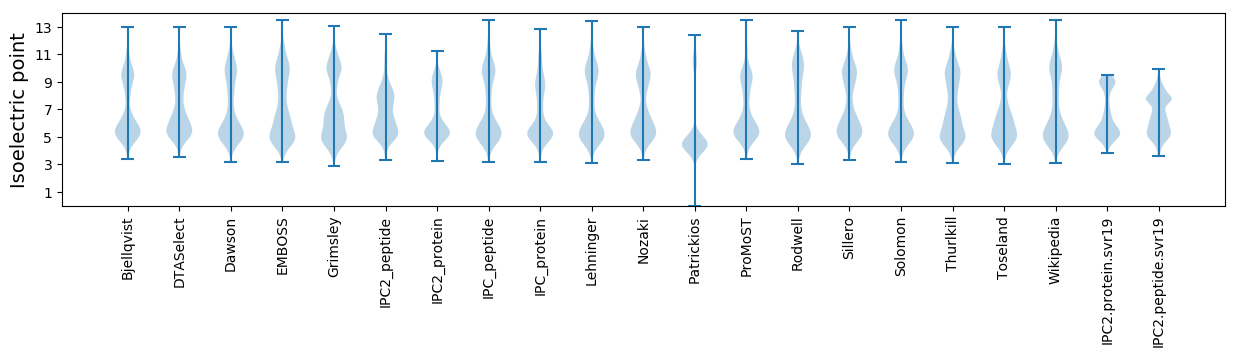
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G7ZH44|A0A6G7ZH44_9SPHN Glycine cleavage system T protein OS=Sphingomonas sp. HDW15A OX=2714942 GN=gcvT PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
649559 |
41 |
1809 |
291.8 |
31.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.77 ± 0.081 | 0.78 ± 0.016 |
5.9 ± 0.037 | 5.966 ± 0.051 |
3.546 ± 0.037 | 8.812 ± 0.064 |
1.881 ± 0.024 | 5.14 ± 0.039 |
3.481 ± 0.042 | 9.924 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.391 ± 0.026 | 2.607 ± 0.036 |
5.18 ± 0.037 | 3.004 ± 0.028 |
7.354 ± 0.062 | 5.559 ± 0.039 |
5.018 ± 0.049 | 7.141 ± 0.047 |
1.411 ± 0.026 | 2.137 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
