
Lacipirellula limnantheis
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Lacipirellulaceae; Lacipirellula
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
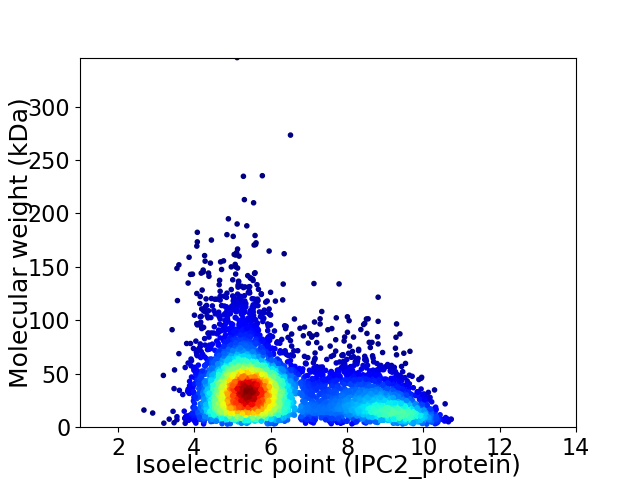
Virtual 2D-PAGE plot for 5493 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A517U649|A0A517U649_9BACT Uncharacterized protein OS=Lacipirellula limnantheis OX=2528024 GN=I41_53530 PE=4 SV=1
MM1 pKa = 8.15DD2 pKa = 5.33KK3 pKa = 10.79YY4 pKa = 11.18QPLRR8 pKa = 11.84DD9 pKa = 3.72ADD11 pKa = 4.3TNGANYY17 pKa = 10.57DD18 pKa = 3.79LDD20 pKa = 4.92ADD22 pKa = 4.04QIIAQLQTWDD32 pKa = 3.19AKK34 pKa = 11.17YY35 pKa = 10.71GVTLSDD41 pKa = 3.54VAHH44 pKa = 7.08DD45 pKa = 3.7AVTVTFNAIPVDD57 pKa = 4.13DD58 pKa = 4.7VPALAAEE65 pKa = 4.49IYY67 pKa = 8.66EE68 pKa = 4.27FCPDD72 pKa = 4.13TIDD75 pKa = 3.45QHH77 pKa = 6.46FGCFAEE83 pKa = 4.62MIEE86 pKa = 4.44MADD89 pKa = 3.48EE90 pKa = 4.49TGEE93 pKa = 4.04EE94 pKa = 4.49LPPEE98 pKa = 4.34LLEE101 pKa = 4.3LTTGVDD107 pKa = 5.24FEE109 pKa = 4.72DD110 pKa = 3.62EE111 pKa = 4.37QYY113 pKa = 11.64GLEE116 pKa = 3.96LLQRR120 pKa = 11.84SLAKK124 pKa = 10.06HH125 pKa = 5.34RR126 pKa = 11.84QVALWWDD133 pKa = 3.36
MM1 pKa = 8.15DD2 pKa = 5.33KK3 pKa = 10.79YY4 pKa = 11.18QPLRR8 pKa = 11.84DD9 pKa = 3.72ADD11 pKa = 4.3TNGANYY17 pKa = 10.57DD18 pKa = 3.79LDD20 pKa = 4.92ADD22 pKa = 4.04QIIAQLQTWDD32 pKa = 3.19AKK34 pKa = 11.17YY35 pKa = 10.71GVTLSDD41 pKa = 3.54VAHH44 pKa = 7.08DD45 pKa = 3.7AVTVTFNAIPVDD57 pKa = 4.13DD58 pKa = 4.7VPALAAEE65 pKa = 4.49IYY67 pKa = 8.66EE68 pKa = 4.27FCPDD72 pKa = 4.13TIDD75 pKa = 3.45QHH77 pKa = 6.46FGCFAEE83 pKa = 4.62MIEE86 pKa = 4.44MADD89 pKa = 3.48EE90 pKa = 4.49TGEE93 pKa = 4.04EE94 pKa = 4.49LPPEE98 pKa = 4.34LLEE101 pKa = 4.3LTTGVDD107 pKa = 5.24FEE109 pKa = 4.72DD110 pKa = 3.62EE111 pKa = 4.37QYY113 pKa = 11.64GLEE116 pKa = 3.96LLQRR120 pKa = 11.84SLAKK124 pKa = 10.06HH125 pKa = 5.34RR126 pKa = 11.84QVALWWDD133 pKa = 3.36
Molecular weight: 15.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A517TYJ6|A0A517TYJ6_9BACT Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Lacipirellula limnantheis OX=2528024 GN=I41_26310 PE=3 SV=1
MM1 pKa = 7.46YY2 pKa = 10.46SPTCHH7 pKa = 6.37SSRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84VLGSSKK18 pKa = 10.6ACHH21 pKa = 5.74QPVEE25 pKa = 4.06LLARR29 pKa = 11.84RR30 pKa = 11.84VVNGARR36 pKa = 11.84IVQRR40 pKa = 11.84PGSSYY45 pKa = 10.76HH46 pKa = 6.32HH47 pKa = 7.2APLPADD53 pKa = 3.44SRR55 pKa = 11.84PVGRR59 pKa = 11.84LALPKK64 pKa = 10.35GDD66 pKa = 3.49TAIDD70 pKa = 3.51QEE72 pKa = 4.34EE73 pKa = 4.3AVAVMVEE80 pKa = 4.29KK81 pKa = 10.43AASISTFEE89 pKa = 4.12GKK91 pKa = 9.77VRR93 pKa = 11.84LRR95 pKa = 11.84WYY97 pKa = 10.66VKK99 pKa = 10.32LQTPEE104 pKa = 3.44ARR106 pKa = 11.84IMLRR110 pKa = 11.84CLRR113 pKa = 11.84LTAAFTCIMLCSVFIVFWMRR133 pKa = 11.84SYY135 pKa = 11.04KK136 pKa = 10.32VQDD139 pKa = 3.88GVTVWYY145 pKa = 8.38ATGKK149 pKa = 7.88TVAFGRR155 pKa = 11.84NEE157 pKa = 3.62QRR159 pKa = 11.84PLISRR164 pKa = 11.84VIVGSTNGTAAVRR177 pKa = 11.84WAAEE181 pKa = 4.3HH182 pKa = 6.12VFSGTPEE189 pKa = 3.81RR190 pKa = 11.84VRR192 pKa = 11.84VEE194 pKa = 3.82FLRR197 pKa = 11.84DD198 pKa = 3.19GGYY201 pKa = 10.73DD202 pKa = 3.57SQWAFKK208 pKa = 10.01KK209 pKa = 10.5RR210 pKa = 11.84SGFAFSEE217 pKa = 4.13NNEE220 pKa = 3.97RR221 pKa = 11.84AVQVAAPYY229 pKa = 9.95WFVTLVTGTAGTLLWMNRR247 pKa = 11.84PYY249 pKa = 11.06RR250 pKa = 11.84FTLRR254 pKa = 11.84GSLAVTTLIALVLGASVVFSRR275 pKa = 5.25
MM1 pKa = 7.46YY2 pKa = 10.46SPTCHH7 pKa = 6.37SSRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84VLGSSKK18 pKa = 10.6ACHH21 pKa = 5.74QPVEE25 pKa = 4.06LLARR29 pKa = 11.84RR30 pKa = 11.84VVNGARR36 pKa = 11.84IVQRR40 pKa = 11.84PGSSYY45 pKa = 10.76HH46 pKa = 6.32HH47 pKa = 7.2APLPADD53 pKa = 3.44SRR55 pKa = 11.84PVGRR59 pKa = 11.84LALPKK64 pKa = 10.35GDD66 pKa = 3.49TAIDD70 pKa = 3.51QEE72 pKa = 4.34EE73 pKa = 4.3AVAVMVEE80 pKa = 4.29KK81 pKa = 10.43AASISTFEE89 pKa = 4.12GKK91 pKa = 9.77VRR93 pKa = 11.84LRR95 pKa = 11.84WYY97 pKa = 10.66VKK99 pKa = 10.32LQTPEE104 pKa = 3.44ARR106 pKa = 11.84IMLRR110 pKa = 11.84CLRR113 pKa = 11.84LTAAFTCIMLCSVFIVFWMRR133 pKa = 11.84SYY135 pKa = 11.04KK136 pKa = 10.32VQDD139 pKa = 3.88GVTVWYY145 pKa = 8.38ATGKK149 pKa = 7.88TVAFGRR155 pKa = 11.84NEE157 pKa = 3.62QRR159 pKa = 11.84PLISRR164 pKa = 11.84VIVGSTNGTAAVRR177 pKa = 11.84WAAEE181 pKa = 4.3HH182 pKa = 6.12VFSGTPEE189 pKa = 3.81RR190 pKa = 11.84VRR192 pKa = 11.84VEE194 pKa = 3.82FLRR197 pKa = 11.84DD198 pKa = 3.19GGYY201 pKa = 10.73DD202 pKa = 3.57SQWAFKK208 pKa = 10.01KK209 pKa = 10.5RR210 pKa = 11.84SGFAFSEE217 pKa = 4.13NNEE220 pKa = 3.97RR221 pKa = 11.84AVQVAAPYY229 pKa = 9.95WFVTLVTGTAGTLLWMNRR247 pKa = 11.84PYY249 pKa = 11.06RR250 pKa = 11.84FTLRR254 pKa = 11.84GSLAVTTLIALVLGASVVFSRR275 pKa = 5.25
Molecular weight: 30.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1881987 |
29 |
3101 |
342.6 |
37.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.005 ± 0.044 | 1.065 ± 0.016 |
5.747 ± 0.026 | 5.864 ± 0.038 |
3.631 ± 0.02 | 8.381 ± 0.051 |
2.038 ± 0.015 | 4.531 ± 0.024 |
3.144 ± 0.031 | 9.871 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.087 ± 0.015 | 3.078 ± 0.03 |
5.431 ± 0.028 | 3.751 ± 0.025 |
6.833 ± 0.033 | 5.927 ± 0.03 |
5.39 ± 0.033 | 7.285 ± 0.026 |
1.578 ± 0.016 | 2.365 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
