
Aeropyrum pernix spindle-shaped virus 1
Taxonomy: Viruses; unclassified archaeal viruses
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
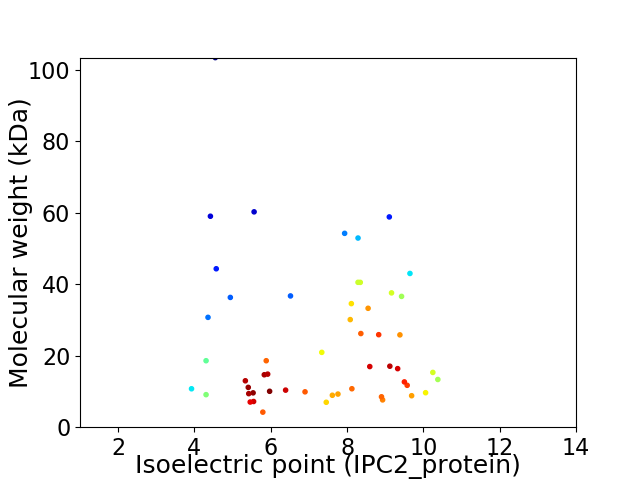
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
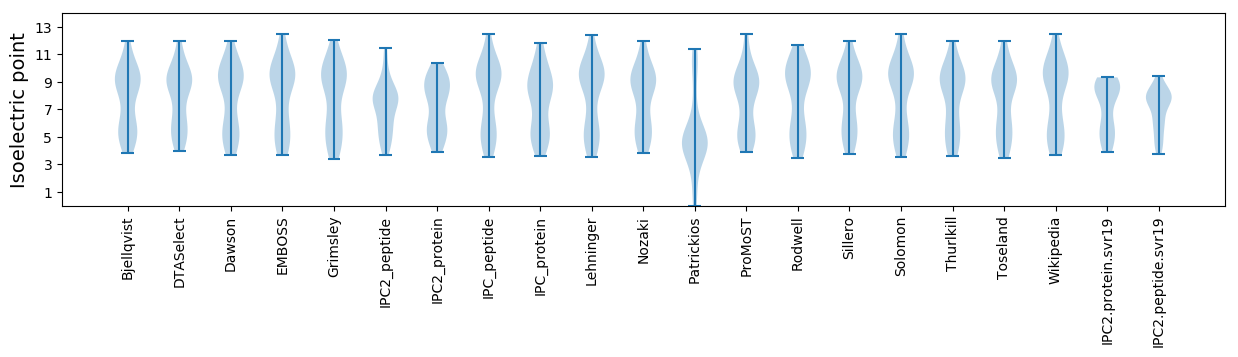
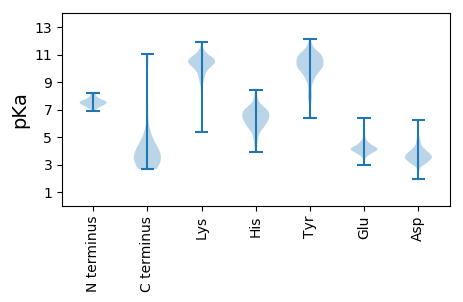
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G3CAY6|G3CAY6_9VIRU Uncharacterized protein OS=Aeropyrum pernix spindle-shaped virus 1 OX=1032473 PE=4 SV=1
MM1 pKa = 7.3QGFTAYY7 pKa = 10.44LVGFAIVAGFAVALGFIFGIFNQIVAQLNTDD38 pKa = 3.81PNNPMISQEE47 pKa = 4.08FVDD50 pKa = 6.1AINLAQSLGGTGLTLAIAALIVGLAMLLIAVIRR83 pKa = 11.84SYY85 pKa = 11.52SGVV88 pKa = 3.23
MM1 pKa = 7.3QGFTAYY7 pKa = 10.44LVGFAIVAGFAVALGFIFGIFNQIVAQLNTDD38 pKa = 3.81PNNPMISQEE47 pKa = 4.08FVDD50 pKa = 6.1AINLAQSLGGTGLTLAIAALIVGLAMLLIAVIRR83 pKa = 11.84SYY85 pKa = 11.52SGVV88 pKa = 3.23
Molecular weight: 9.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G3CAY0|G3CAY0_9VIRU Uncharacterized protein OS=Aeropyrum pernix spindle-shaped virus 1 OX=1032473 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 5.04GGGSAGSAPSSPGSSRR18 pKa = 11.84PPDD21 pKa = 3.34YY22 pKa = 11.03SLYY25 pKa = 10.34EE26 pKa = 4.16SQPRR30 pKa = 11.84PAPASWSPLGVLASALDD47 pKa = 3.6APYY50 pKa = 11.09SFVQEE55 pKa = 4.27RR56 pKa = 11.84GWGVVEE62 pKa = 4.06RR63 pKa = 11.84LRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84ALRR70 pKa = 11.84EE71 pKa = 3.79KK72 pKa = 11.22AEE74 pKa = 3.9GDD76 pKa = 3.74YY77 pKa = 11.74AEE79 pKa = 4.61GFLWALGASAASFGVGVAAGVTGLFSPRR107 pKa = 11.84AWRR110 pKa = 11.84EE111 pKa = 3.48AFHH114 pKa = 8.07AFMHH118 pKa = 6.42PGEE121 pKa = 4.35TVRR124 pKa = 11.84AVASDD129 pKa = 3.94PLSWPYY135 pKa = 10.76LAGSVVGPAKK145 pKa = 10.52LGRR148 pKa = 11.84AVGRR152 pKa = 11.84RR153 pKa = 11.84LGRR156 pKa = 11.84VRR158 pKa = 11.84EE159 pKa = 4.06PDD161 pKa = 3.27LASTSEE167 pKa = 4.04GSLTLVAAKK176 pKa = 10.28GEE178 pKa = 4.14RR179 pKa = 11.84LRR181 pKa = 11.84WAARR185 pKa = 11.84LEE187 pKa = 4.08ARR189 pKa = 11.84GPTVGVKK196 pKa = 9.63GVKK199 pKa = 9.24PRR201 pKa = 11.84PPPDD205 pKa = 3.1RR206 pKa = 11.84LVTLEE211 pKa = 4.04TPRR214 pKa = 11.84GRR216 pKa = 11.84VEE218 pKa = 3.91LLEE221 pKa = 4.09SRR223 pKa = 11.84RR224 pKa = 11.84GSTVRR229 pKa = 11.84RR230 pKa = 11.84VYY232 pKa = 9.91RR233 pKa = 11.84AEE235 pKa = 3.71YY236 pKa = 10.09RR237 pKa = 11.84DD238 pKa = 3.59VGKK241 pKa = 10.75KK242 pKa = 10.29LLGVEE247 pKa = 4.27EE248 pKa = 4.51WSVEE252 pKa = 3.28GWLRR256 pKa = 11.84PRR258 pKa = 11.84YY259 pKa = 9.28RR260 pKa = 11.84YY261 pKa = 9.79RR262 pKa = 11.84GLLVDD267 pKa = 4.64PEE269 pKa = 4.65TPGAAVLLEE278 pKa = 3.79ARR280 pKa = 11.84RR281 pKa = 11.84PLLRR285 pKa = 11.84GVDD288 pKa = 4.22LPPPRR293 pKa = 11.84LWVEE297 pKa = 3.72PRR299 pKa = 11.84LAAPAGAVGLILGLGAAGTGARR321 pKa = 11.84GGLEE325 pKa = 3.8GGAAGRR331 pKa = 11.84PAEE334 pKa = 4.33AGARR338 pKa = 11.84GLVSGSSLAGGEE350 pKa = 4.2AAASTLAGASAASSPAEE367 pKa = 3.96PSAASAGRR375 pKa = 11.84GGPEE379 pKa = 3.92APPLLGSGSRR389 pKa = 11.84GRR391 pKa = 11.84RR392 pKa = 11.84GRR394 pKa = 11.84RR395 pKa = 11.84GKK397 pKa = 10.02LAYY400 pKa = 10.23SEE402 pKa = 4.01ILYY405 pKa = 10.02PGGVLLEE412 pKa = 4.25LLL414 pKa = 4.31
MM1 pKa = 8.12DD2 pKa = 5.04GGGSAGSAPSSPGSSRR18 pKa = 11.84PPDD21 pKa = 3.34YY22 pKa = 11.03SLYY25 pKa = 10.34EE26 pKa = 4.16SQPRR30 pKa = 11.84PAPASWSPLGVLASALDD47 pKa = 3.6APYY50 pKa = 11.09SFVQEE55 pKa = 4.27RR56 pKa = 11.84GWGVVEE62 pKa = 4.06RR63 pKa = 11.84LRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84ALRR70 pKa = 11.84EE71 pKa = 3.79KK72 pKa = 11.22AEE74 pKa = 3.9GDD76 pKa = 3.74YY77 pKa = 11.74AEE79 pKa = 4.61GFLWALGASAASFGVGVAAGVTGLFSPRR107 pKa = 11.84AWRR110 pKa = 11.84EE111 pKa = 3.48AFHH114 pKa = 8.07AFMHH118 pKa = 6.42PGEE121 pKa = 4.35TVRR124 pKa = 11.84AVASDD129 pKa = 3.94PLSWPYY135 pKa = 10.76LAGSVVGPAKK145 pKa = 10.52LGRR148 pKa = 11.84AVGRR152 pKa = 11.84RR153 pKa = 11.84LGRR156 pKa = 11.84VRR158 pKa = 11.84EE159 pKa = 4.06PDD161 pKa = 3.27LASTSEE167 pKa = 4.04GSLTLVAAKK176 pKa = 10.28GEE178 pKa = 4.14RR179 pKa = 11.84LRR181 pKa = 11.84WAARR185 pKa = 11.84LEE187 pKa = 4.08ARR189 pKa = 11.84GPTVGVKK196 pKa = 9.63GVKK199 pKa = 9.24PRR201 pKa = 11.84PPPDD205 pKa = 3.1RR206 pKa = 11.84LVTLEE211 pKa = 4.04TPRR214 pKa = 11.84GRR216 pKa = 11.84VEE218 pKa = 3.91LLEE221 pKa = 4.09SRR223 pKa = 11.84RR224 pKa = 11.84GSTVRR229 pKa = 11.84RR230 pKa = 11.84VYY232 pKa = 9.91RR233 pKa = 11.84AEE235 pKa = 3.71YY236 pKa = 10.09RR237 pKa = 11.84DD238 pKa = 3.59VGKK241 pKa = 10.75KK242 pKa = 10.29LLGVEE247 pKa = 4.27EE248 pKa = 4.51WSVEE252 pKa = 3.28GWLRR256 pKa = 11.84PRR258 pKa = 11.84YY259 pKa = 9.28RR260 pKa = 11.84YY261 pKa = 9.79RR262 pKa = 11.84GLLVDD267 pKa = 4.64PEE269 pKa = 4.65TPGAAVLLEE278 pKa = 3.79ARR280 pKa = 11.84RR281 pKa = 11.84PLLRR285 pKa = 11.84GVDD288 pKa = 4.22LPPPRR293 pKa = 11.84LWVEE297 pKa = 3.72PRR299 pKa = 11.84LAAPAGAVGLILGLGAAGTGARR321 pKa = 11.84GGLEE325 pKa = 3.8GGAAGRR331 pKa = 11.84PAEE334 pKa = 4.33AGARR338 pKa = 11.84GLVSGSSLAGGEE350 pKa = 4.2AAASTLAGASAASSPAEE367 pKa = 3.96PSAASAGRR375 pKa = 11.84GGPEE379 pKa = 3.92APPLLGSGSRR389 pKa = 11.84GRR391 pKa = 11.84RR392 pKa = 11.84GRR394 pKa = 11.84RR395 pKa = 11.84GKK397 pKa = 10.02LAYY400 pKa = 10.23SEE402 pKa = 4.01ILYY405 pKa = 10.02PGGVLLEE412 pKa = 4.25LLL414 pKa = 4.31
Molecular weight: 43.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11486 |
38 |
929 |
216.7 |
24.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.324 ± 0.421 | 0.609 ± 0.095 |
4.788 ± 0.239 | 7.191 ± 0.309 |
3.213 ± 0.209 | 8.375 ± 0.518 |
1.41 ± 0.134 | 4.78 ± 0.495 |
4.083 ± 0.428 | 11.675 ± 0.295 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.037 ± 0.147 | 2.046 ± 0.196 |
4.536 ± 0.293 | 2.168 ± 0.22 |
8.593 ± 0.582 | 6.26 ± 0.253 |
4.066 ± 0.286 | 8.523 ± 0.358 |
1.898 ± 0.214 | 4.423 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
