
Citrus leprosis virus C2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Kitaviridae; Cilevirus
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
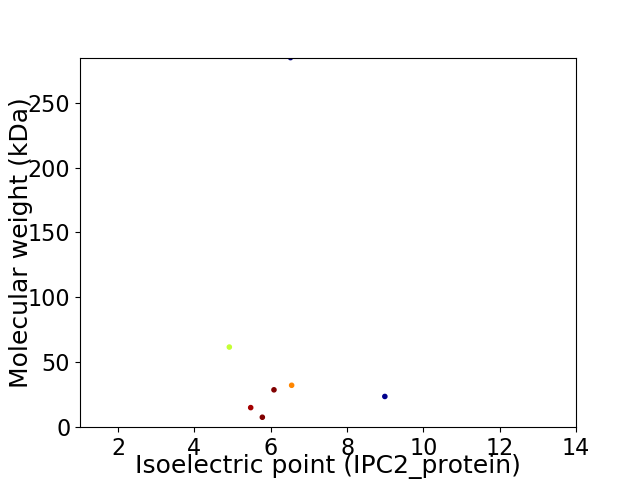
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1JAW3|M1JAW3_9VIRU Helicase OS=Citrus leprosis virus C2 OX=2052685 PE=4 SV=1
MM1 pKa = 8.06DD2 pKa = 5.45FYY4 pKa = 11.13TVYY7 pKa = 10.65TFLNLTLGLLTYY19 pKa = 10.53NYY21 pKa = 10.39DD22 pKa = 3.47SLGTFGPTPRR32 pKa = 11.84CTSKK36 pKa = 10.7RR37 pKa = 11.84CSLLPAEE44 pKa = 5.59CSTEE48 pKa = 3.82HH49 pKa = 5.69TVNKK53 pKa = 9.67VVEE56 pKa = 4.26RR57 pKa = 11.84HH58 pKa = 5.0VFTANDD64 pKa = 4.09HH65 pKa = 5.74YY66 pKa = 10.79CISLGYY72 pKa = 9.53RR73 pKa = 11.84KK74 pKa = 10.18NISFSVSTLFIGDD87 pKa = 3.48YY88 pKa = 10.48DD89 pKa = 4.04VYY91 pKa = 11.44GGFEE95 pKa = 4.45CDD97 pKa = 2.88RR98 pKa = 11.84TALSMGIVSNRR109 pKa = 11.84LGFYY113 pKa = 10.5EE114 pKa = 5.77DD115 pKa = 5.2ALFDD119 pKa = 4.01CKK121 pKa = 10.89NYY123 pKa = 10.05NLSYY127 pKa = 10.76GNYY130 pKa = 9.39RR131 pKa = 11.84FEE133 pKa = 4.3FCLNSSVYY141 pKa = 10.92DD142 pKa = 3.53SVLTDD147 pKa = 3.63KK148 pKa = 10.98FYY150 pKa = 10.91TPQIEE155 pKa = 3.93AAAPINTRR163 pKa = 11.84GVEE166 pKa = 4.08VFDD169 pKa = 4.95DD170 pKa = 4.39FLGFTYY176 pKa = 10.71YY177 pKa = 9.17RR178 pKa = 11.84TSCDD182 pKa = 3.56SFCSEE187 pKa = 4.97SKK189 pKa = 10.65FSPDD193 pKa = 2.99FDD195 pKa = 3.94FGNYY199 pKa = 7.4VFYY202 pKa = 10.68FPLCDD207 pKa = 3.38NHH209 pKa = 7.87IPLCYY214 pKa = 10.3DD215 pKa = 3.39GAEE218 pKa = 4.22DD219 pKa = 3.76SCPRR223 pKa = 11.84GYY225 pKa = 10.78SLQTIFVSSLVEE237 pKa = 4.0EE238 pKa = 4.49EE239 pKa = 4.37TEE241 pKa = 3.74KK242 pKa = 10.66LARR245 pKa = 11.84VICVSDD251 pKa = 3.89NEE253 pKa = 4.46PGLTFPAWFRR263 pKa = 11.84DD264 pKa = 3.74NPSTYY269 pKa = 10.96DD270 pKa = 3.82DD271 pKa = 4.25LVGNYY276 pKa = 7.67TINVEE281 pKa = 4.59SKK283 pKa = 10.65CGVAMSKK290 pKa = 10.58KK291 pKa = 9.43MRR293 pKa = 11.84FSRR296 pKa = 11.84YY297 pKa = 8.96GVTGTYY303 pKa = 9.32IDD305 pKa = 5.17DD306 pKa = 4.71IIVVTIDD313 pKa = 3.29EE314 pKa = 4.51VHH316 pKa = 6.32FLTSKK321 pKa = 10.09FCRR324 pKa = 11.84DD325 pKa = 3.56YY326 pKa = 11.37ILFLGYY332 pKa = 9.85RR333 pKa = 11.84DD334 pKa = 4.94GYY336 pKa = 11.62DD337 pKa = 3.18NDD339 pKa = 4.58FSMKK343 pKa = 10.04TYY345 pKa = 9.04FQKK348 pKa = 10.87RR349 pKa = 11.84LNCNSDD355 pKa = 3.39GCWYY359 pKa = 10.75SGVDD363 pKa = 3.34LSRR366 pKa = 11.84ILSHH370 pKa = 7.07CEE372 pKa = 3.12LSLVVEE378 pKa = 4.46KK379 pKa = 11.08KK380 pKa = 9.64EE381 pKa = 3.93ALVSTFTLFNKK392 pKa = 9.33TFGGKK397 pKa = 9.54VGFIPVGLSDD407 pKa = 4.45SVLFGYY413 pKa = 10.51NVFLFRR419 pKa = 11.84GFYY422 pKa = 9.92SYY424 pKa = 11.33SSSSVTLKK432 pKa = 8.39STKK435 pKa = 10.09YY436 pKa = 10.05FLVKK440 pKa = 10.58AEE442 pKa = 4.49AEE444 pKa = 4.2WYY446 pKa = 7.22MKK448 pKa = 11.21LMMFFADD455 pKa = 5.39DD456 pKa = 3.55VLKK459 pKa = 11.25VCFEE463 pKa = 4.73TIFSVLLGALSSCLSFIFNIGGCCFRR489 pKa = 11.84LVFLCVMDD497 pKa = 4.69SIIILLCLLPCYY509 pKa = 9.95CHH511 pKa = 7.28LGFILCSVSNLIIKK525 pKa = 10.47LFMKK529 pKa = 10.24NNCCFGISDD538 pKa = 4.25AVASSFF544 pKa = 4.31
MM1 pKa = 8.06DD2 pKa = 5.45FYY4 pKa = 11.13TVYY7 pKa = 10.65TFLNLTLGLLTYY19 pKa = 10.53NYY21 pKa = 10.39DD22 pKa = 3.47SLGTFGPTPRR32 pKa = 11.84CTSKK36 pKa = 10.7RR37 pKa = 11.84CSLLPAEE44 pKa = 5.59CSTEE48 pKa = 3.82HH49 pKa = 5.69TVNKK53 pKa = 9.67VVEE56 pKa = 4.26RR57 pKa = 11.84HH58 pKa = 5.0VFTANDD64 pKa = 4.09HH65 pKa = 5.74YY66 pKa = 10.79CISLGYY72 pKa = 9.53RR73 pKa = 11.84KK74 pKa = 10.18NISFSVSTLFIGDD87 pKa = 3.48YY88 pKa = 10.48DD89 pKa = 4.04VYY91 pKa = 11.44GGFEE95 pKa = 4.45CDD97 pKa = 2.88RR98 pKa = 11.84TALSMGIVSNRR109 pKa = 11.84LGFYY113 pKa = 10.5EE114 pKa = 5.77DD115 pKa = 5.2ALFDD119 pKa = 4.01CKK121 pKa = 10.89NYY123 pKa = 10.05NLSYY127 pKa = 10.76GNYY130 pKa = 9.39RR131 pKa = 11.84FEE133 pKa = 4.3FCLNSSVYY141 pKa = 10.92DD142 pKa = 3.53SVLTDD147 pKa = 3.63KK148 pKa = 10.98FYY150 pKa = 10.91TPQIEE155 pKa = 3.93AAAPINTRR163 pKa = 11.84GVEE166 pKa = 4.08VFDD169 pKa = 4.95DD170 pKa = 4.39FLGFTYY176 pKa = 10.71YY177 pKa = 9.17RR178 pKa = 11.84TSCDD182 pKa = 3.56SFCSEE187 pKa = 4.97SKK189 pKa = 10.65FSPDD193 pKa = 2.99FDD195 pKa = 3.94FGNYY199 pKa = 7.4VFYY202 pKa = 10.68FPLCDD207 pKa = 3.38NHH209 pKa = 7.87IPLCYY214 pKa = 10.3DD215 pKa = 3.39GAEE218 pKa = 4.22DD219 pKa = 3.76SCPRR223 pKa = 11.84GYY225 pKa = 10.78SLQTIFVSSLVEE237 pKa = 4.0EE238 pKa = 4.49EE239 pKa = 4.37TEE241 pKa = 3.74KK242 pKa = 10.66LARR245 pKa = 11.84VICVSDD251 pKa = 3.89NEE253 pKa = 4.46PGLTFPAWFRR263 pKa = 11.84DD264 pKa = 3.74NPSTYY269 pKa = 10.96DD270 pKa = 3.82DD271 pKa = 4.25LVGNYY276 pKa = 7.67TINVEE281 pKa = 4.59SKK283 pKa = 10.65CGVAMSKK290 pKa = 10.58KK291 pKa = 9.43MRR293 pKa = 11.84FSRR296 pKa = 11.84YY297 pKa = 8.96GVTGTYY303 pKa = 9.32IDD305 pKa = 5.17DD306 pKa = 4.71IIVVTIDD313 pKa = 3.29EE314 pKa = 4.51VHH316 pKa = 6.32FLTSKK321 pKa = 10.09FCRR324 pKa = 11.84DD325 pKa = 3.56YY326 pKa = 11.37ILFLGYY332 pKa = 9.85RR333 pKa = 11.84DD334 pKa = 4.94GYY336 pKa = 11.62DD337 pKa = 3.18NDD339 pKa = 4.58FSMKK343 pKa = 10.04TYY345 pKa = 9.04FQKK348 pKa = 10.87RR349 pKa = 11.84LNCNSDD355 pKa = 3.39GCWYY359 pKa = 10.75SGVDD363 pKa = 3.34LSRR366 pKa = 11.84ILSHH370 pKa = 7.07CEE372 pKa = 3.12LSLVVEE378 pKa = 4.46KK379 pKa = 11.08KK380 pKa = 9.64EE381 pKa = 3.93ALVSTFTLFNKK392 pKa = 9.33TFGGKK397 pKa = 9.54VGFIPVGLSDD407 pKa = 4.45SVLFGYY413 pKa = 10.51NVFLFRR419 pKa = 11.84GFYY422 pKa = 9.92SYY424 pKa = 11.33SSSSVTLKK432 pKa = 8.39STKK435 pKa = 10.09YY436 pKa = 10.05FLVKK440 pKa = 10.58AEE442 pKa = 4.49AEE444 pKa = 4.2WYY446 pKa = 7.22MKK448 pKa = 11.21LMMFFADD455 pKa = 5.39DD456 pKa = 3.55VLKK459 pKa = 11.25VCFEE463 pKa = 4.73TIFSVLLGALSSCLSFIFNIGGCCFRR489 pKa = 11.84LVFLCVMDD497 pKa = 4.69SIIILLCLLPCYY509 pKa = 9.95CHH511 pKa = 7.28LGFILCSVSNLIIKK525 pKa = 10.47LFMKK529 pKa = 10.24NNCCFGISDD538 pKa = 4.25AVASSFF544 pKa = 4.31
Molecular weight: 61.74 kDa
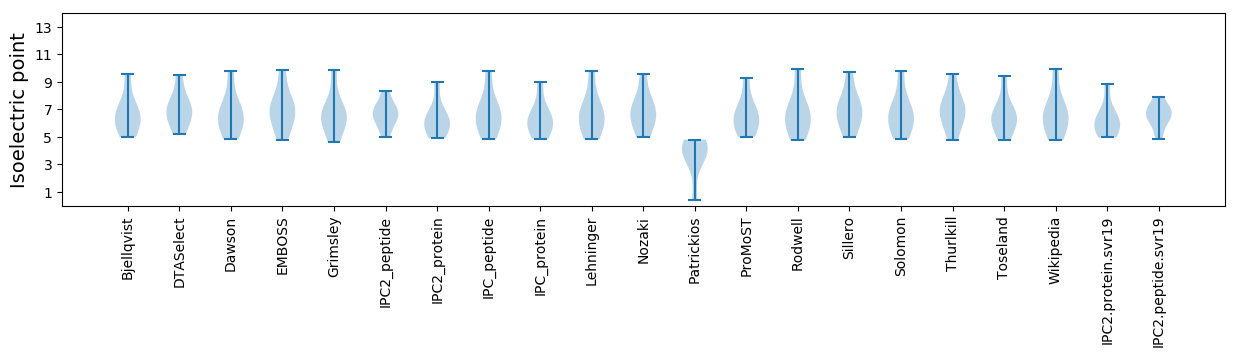
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1J7K9|M1J7K9_9VIRU p61 OS=Citrus leprosis virus C2 OX=2052685 PE=4 SV=1
MM1 pKa = 7.77DD2 pKa = 3.27QRR4 pKa = 11.84FLRR7 pKa = 11.84GASVLNLSNKK17 pKa = 9.48RR18 pKa = 11.84EE19 pKa = 4.01KK20 pKa = 10.98LKK22 pKa = 10.42TNASVSLLSDD32 pKa = 3.53IQQILLRR39 pKa = 11.84YY40 pKa = 7.55IQKK43 pKa = 10.39PYY45 pKa = 10.02VLLMYY50 pKa = 10.67ACVLVLFAMHH60 pKa = 6.85IDD62 pKa = 3.84AGTHH66 pKa = 7.5DD67 pKa = 4.75ILDD70 pKa = 4.44DD71 pKa = 4.72LAQQFPNNPVIEE83 pKa = 4.18WARR86 pKa = 11.84NNFFRR91 pKa = 11.84LCGALVFIPVITDD104 pKa = 2.94ARR106 pKa = 11.84KK107 pKa = 8.45EE108 pKa = 3.91HH109 pKa = 5.76QLYY112 pKa = 10.33FGMVIALFLLGFPQRR127 pKa = 11.84SIFEE131 pKa = 4.19YY132 pKa = 10.2FVYY135 pKa = 10.57SLSLHH140 pKa = 6.29VYY142 pKa = 9.79AKK144 pKa = 10.14SKK146 pKa = 11.06HH147 pKa = 5.14PFTRR151 pKa = 11.84IFILVITVVSCVLFGVFTNDD171 pKa = 3.36QLRR174 pKa = 11.84KK175 pKa = 9.91LYY177 pKa = 10.47QEE179 pKa = 4.18LPKK182 pKa = 10.78VPTHH186 pKa = 6.25PVNKK190 pKa = 9.71VEE192 pKa = 4.09RR193 pKa = 11.84VVNRR197 pKa = 11.84VGGQQSFSGG206 pKa = 3.64
MM1 pKa = 7.77DD2 pKa = 3.27QRR4 pKa = 11.84FLRR7 pKa = 11.84GASVLNLSNKK17 pKa = 9.48RR18 pKa = 11.84EE19 pKa = 4.01KK20 pKa = 10.98LKK22 pKa = 10.42TNASVSLLSDD32 pKa = 3.53IQQILLRR39 pKa = 11.84YY40 pKa = 7.55IQKK43 pKa = 10.39PYY45 pKa = 10.02VLLMYY50 pKa = 10.67ACVLVLFAMHH60 pKa = 6.85IDD62 pKa = 3.84AGTHH66 pKa = 7.5DD67 pKa = 4.75ILDD70 pKa = 4.44DD71 pKa = 4.72LAQQFPNNPVIEE83 pKa = 4.18WARR86 pKa = 11.84NNFFRR91 pKa = 11.84LCGALVFIPVITDD104 pKa = 2.94ARR106 pKa = 11.84KK107 pKa = 8.45EE108 pKa = 3.91HH109 pKa = 5.76QLYY112 pKa = 10.33FGMVIALFLLGFPQRR127 pKa = 11.84SIFEE131 pKa = 4.19YY132 pKa = 10.2FVYY135 pKa = 10.57SLSLHH140 pKa = 6.29VYY142 pKa = 9.79AKK144 pKa = 10.14SKK146 pKa = 11.06HH147 pKa = 5.14PFTRR151 pKa = 11.84IFILVITVVSCVLFGVFTNDD171 pKa = 3.36QLRR174 pKa = 11.84KK175 pKa = 9.91LYY177 pKa = 10.47QEE179 pKa = 4.18LPKK182 pKa = 10.78VPTHH186 pKa = 6.25PVNKK190 pKa = 9.71VEE192 pKa = 4.09RR193 pKa = 11.84VVNRR197 pKa = 11.84VGGQQSFSGG206 pKa = 3.64
Molecular weight: 23.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4007 |
67 |
2504 |
572.4 |
64.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.091 ± 0.488 | 2.945 ± 0.538 |
6.613 ± 0.597 | 4.842 ± 0.448 |
6.289 ± 0.819 | 5.141 ± 0.5 |
2.595 ± 0.311 | 6.114 ± 0.519 |
5.54 ± 0.755 | 9.733 ± 0.402 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.997 ± 0.286 | 4.817 ± 0.509 |
3.893 ± 1.183 | 2.62 ± 0.627 |
5.49 ± 0.535 | 8.56 ± 0.567 |
5.091 ± 0.42 | 8.111 ± 0.291 |
0.574 ± 0.092 | 3.943 ± 0.486 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
