
Lactobacillus kimchicus JCM 15530
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Secundilactobacillus; Secundilactobacillus kimchicus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
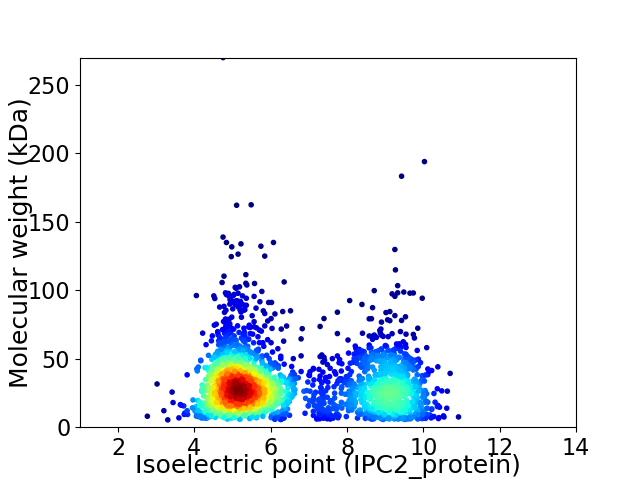
Virtual 2D-PAGE plot for 2470 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R1HWX8|A0A0R1HWX8_9LACO CSD domain-containing protein OS=Lactobacillus kimchicus JCM 15530 OX=1302272 GN=FC96_GL002087 PE=4 SV=1
MM1 pKa = 7.35TFSFLLFSSTFGFTVVGNTQAQATEE26 pKa = 4.17TPTAVVQNVKK36 pKa = 9.78TSTPKK41 pKa = 10.18AHH43 pKa = 6.62ATFTTSVKK51 pKa = 10.55SINQVVQADD60 pKa = 4.28DD61 pKa = 3.69YY62 pKa = 10.91ATAITQEE69 pKa = 4.36VNWINTQVAAEE80 pKa = 4.88KK81 pKa = 9.56LTGWDD86 pKa = 2.74ALALVRR92 pKa = 11.84SSTGITDD99 pKa = 3.52AQKK102 pKa = 8.67TQIQTNIEE110 pKa = 3.99TNYY113 pKa = 10.64ADD115 pKa = 5.49PSTQHH120 pKa = 5.95QPTDD124 pKa = 3.39YY125 pKa = 10.82ARR127 pKa = 11.84DD128 pKa = 3.91VIGLICVGADD138 pKa = 3.24PTNVNGQNLVQEE150 pKa = 4.51AVTNGIAADD159 pKa = 3.62ADD161 pKa = 4.39VYY163 pKa = 11.4AVTYY167 pKa = 10.45GLIAATAAQSSNLGNVQTADD187 pKa = 3.67LQIMVTNLLGMQNSEE202 pKa = 3.66TGYY205 pKa = 8.01WTDD208 pKa = 2.95QYY210 pKa = 11.52GYY212 pKa = 8.34TLDD215 pKa = 3.68TTGMALQALALYY227 pKa = 9.89LGNSEE232 pKa = 4.62FDD234 pKa = 3.07TDD236 pKa = 3.98GRR238 pKa = 11.84ITNAVNSAFNTITLIGGNGALQEE261 pKa = 4.79DD262 pKa = 5.54GNFKK266 pKa = 11.23DD267 pKa = 4.53PFVADD272 pKa = 3.55AAVNSSSDD280 pKa = 3.07AMLIAGLAACGTEE293 pKa = 5.09LPDD296 pKa = 4.23EE297 pKa = 4.85SPLSALLSFQVLHH310 pKa = 6.07GTDD313 pKa = 2.97AGSFNWTPEE322 pKa = 3.75MTYY325 pKa = 10.99DD326 pKa = 3.8RR327 pKa = 11.84QLSTQQAVYY336 pKa = 10.96ALDD339 pKa = 3.5QYY341 pKa = 11.57SYY343 pKa = 11.51SVDD346 pKa = 3.11GKK348 pKa = 11.43GSIFDD353 pKa = 3.65FTANPVNN360 pKa = 3.62
MM1 pKa = 7.35TFSFLLFSSTFGFTVVGNTQAQATEE26 pKa = 4.17TPTAVVQNVKK36 pKa = 9.78TSTPKK41 pKa = 10.18AHH43 pKa = 6.62ATFTTSVKK51 pKa = 10.55SINQVVQADD60 pKa = 4.28DD61 pKa = 3.69YY62 pKa = 10.91ATAITQEE69 pKa = 4.36VNWINTQVAAEE80 pKa = 4.88KK81 pKa = 9.56LTGWDD86 pKa = 2.74ALALVRR92 pKa = 11.84SSTGITDD99 pKa = 3.52AQKK102 pKa = 8.67TQIQTNIEE110 pKa = 3.99TNYY113 pKa = 10.64ADD115 pKa = 5.49PSTQHH120 pKa = 5.95QPTDD124 pKa = 3.39YY125 pKa = 10.82ARR127 pKa = 11.84DD128 pKa = 3.91VIGLICVGADD138 pKa = 3.24PTNVNGQNLVQEE150 pKa = 4.51AVTNGIAADD159 pKa = 3.62ADD161 pKa = 4.39VYY163 pKa = 11.4AVTYY167 pKa = 10.45GLIAATAAQSSNLGNVQTADD187 pKa = 3.67LQIMVTNLLGMQNSEE202 pKa = 3.66TGYY205 pKa = 8.01WTDD208 pKa = 2.95QYY210 pKa = 11.52GYY212 pKa = 8.34TLDD215 pKa = 3.68TTGMALQALALYY227 pKa = 9.89LGNSEE232 pKa = 4.62FDD234 pKa = 3.07TDD236 pKa = 3.98GRR238 pKa = 11.84ITNAVNSAFNTITLIGGNGALQEE261 pKa = 4.79DD262 pKa = 5.54GNFKK266 pKa = 11.23DD267 pKa = 4.53PFVADD272 pKa = 3.55AAVNSSSDD280 pKa = 3.07AMLIAGLAACGTEE293 pKa = 5.09LPDD296 pKa = 4.23EE297 pKa = 4.85SPLSALLSFQVLHH310 pKa = 6.07GTDD313 pKa = 2.97AGSFNWTPEE322 pKa = 3.75MTYY325 pKa = 10.99DD326 pKa = 3.8RR327 pKa = 11.84QLSTQQAVYY336 pKa = 10.96ALDD339 pKa = 3.5QYY341 pKa = 11.57SYY343 pKa = 11.51SVDD346 pKa = 3.11GKK348 pKa = 11.43GSIFDD353 pKa = 3.65FTANPVNN360 pKa = 3.62
Molecular weight: 38.22 kDa
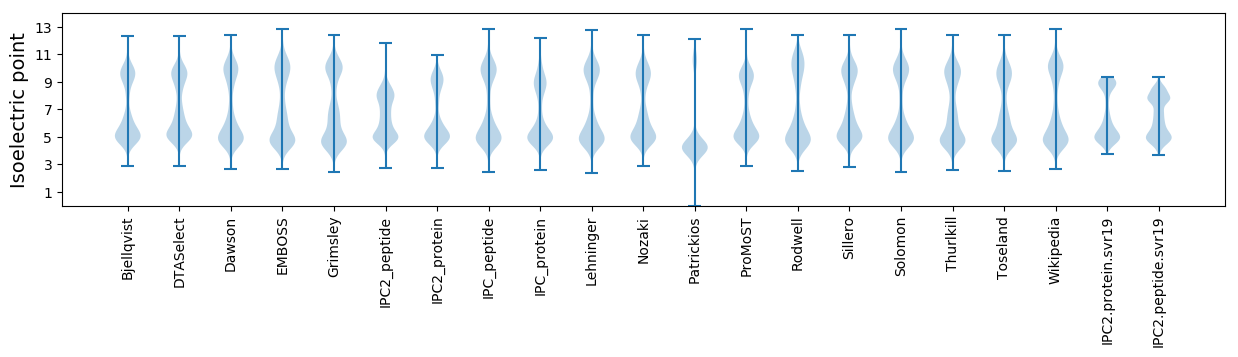
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R1HMB6|A0A0R1HMB6_9LACO HTH arsR-type domain-containing protein OS=Lactobacillus kimchicus JCM 15530 OX=1302272 GN=FC96_GL002213 PE=4 SV=1
MM1 pKa = 7.45VYY3 pKa = 9.73VGKK6 pKa = 10.69RR7 pKa = 11.84IDD9 pKa = 3.54LTGNRR14 pKa = 11.84YY15 pKa = 9.13SRR17 pKa = 11.84LMVVSFVGNSKK28 pKa = 10.47NGNARR33 pKa = 11.84WLCEE37 pKa = 3.95CDD39 pKa = 2.74CGRR42 pKa = 11.84FIEE45 pKa = 4.35TDD47 pKa = 3.72GYY49 pKa = 10.46RR50 pKa = 11.84LRR52 pKa = 11.84TGRR55 pKa = 11.84VRR57 pKa = 11.84SCGCLRR63 pKa = 11.84RR64 pKa = 11.84EE65 pKa = 4.01KK66 pKa = 11.0SRR68 pKa = 11.84DD69 pKa = 3.32RR70 pKa = 11.84LLGFAATKK78 pKa = 10.43QNIGVADD85 pKa = 4.18NLPIHH90 pKa = 6.55NGTVLRR96 pKa = 11.84LTDD99 pKa = 3.86KK100 pKa = 10.92NNQSGVIGVSWDD112 pKa = 3.32KK113 pKa = 11.16QSQKK117 pKa = 8.88WVARR121 pKa = 11.84LMVRR125 pKa = 11.84GKK127 pKa = 10.43YY128 pKa = 9.42VINRR132 pKa = 11.84KK133 pKa = 7.83FHH135 pKa = 6.25NFNQAVSFRR144 pKa = 11.84HH145 pKa = 4.49QAEE148 pKa = 4.12LQFRR152 pKa = 11.84DD153 pKa = 3.39PLLRR157 pKa = 5.42
MM1 pKa = 7.45VYY3 pKa = 9.73VGKK6 pKa = 10.69RR7 pKa = 11.84IDD9 pKa = 3.54LTGNRR14 pKa = 11.84YY15 pKa = 9.13SRR17 pKa = 11.84LMVVSFVGNSKK28 pKa = 10.47NGNARR33 pKa = 11.84WLCEE37 pKa = 3.95CDD39 pKa = 2.74CGRR42 pKa = 11.84FIEE45 pKa = 4.35TDD47 pKa = 3.72GYY49 pKa = 10.46RR50 pKa = 11.84LRR52 pKa = 11.84TGRR55 pKa = 11.84VRR57 pKa = 11.84SCGCLRR63 pKa = 11.84RR64 pKa = 11.84EE65 pKa = 4.01KK66 pKa = 11.0SRR68 pKa = 11.84DD69 pKa = 3.32RR70 pKa = 11.84LLGFAATKK78 pKa = 10.43QNIGVADD85 pKa = 4.18NLPIHH90 pKa = 6.55NGTVLRR96 pKa = 11.84LTDD99 pKa = 3.86KK100 pKa = 10.92NNQSGVIGVSWDD112 pKa = 3.32KK113 pKa = 11.16QSQKK117 pKa = 8.88WVARR121 pKa = 11.84LMVRR125 pKa = 11.84GKK127 pKa = 10.43YY128 pKa = 9.42VINRR132 pKa = 11.84KK133 pKa = 7.83FHH135 pKa = 6.25NFNQAVSFRR144 pKa = 11.84HH145 pKa = 4.49QAEE148 pKa = 4.12LQFRR152 pKa = 11.84DD153 pKa = 3.39PLLRR157 pKa = 5.42
Molecular weight: 18.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
740804 |
50 |
2467 |
299.9 |
33.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.248 ± 0.063 | 0.375 ± 0.01 |
5.822 ± 0.051 | 4.965 ± 0.053 |
4.198 ± 0.042 | 7.122 ± 0.05 |
2.172 ± 0.024 | 6.356 ± 0.045 |
5.357 ± 0.054 | 9.897 ± 0.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.687 ± 0.03 | 4.332 ± 0.035 |
3.876 ± 0.028 | 4.665 ± 0.04 |
4.314 ± 0.036 | 5.861 ± 0.056 |
6.787 ± 0.052 | 7.597 ± 0.044 |
1.082 ± 0.018 | 3.287 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
