
Candidatus Tokpelaia hoelldoblerii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiales incertae sedis; Candidatus Tokpelaia
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
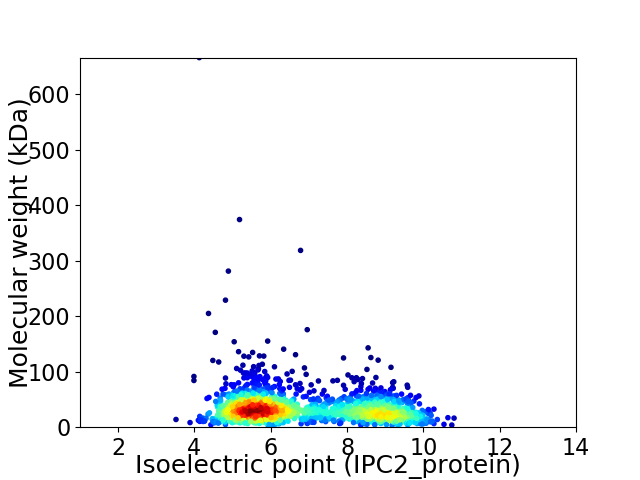
Virtual 2D-PAGE plot for 1683 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U9JSK4|A0A1U9JSK4_9RHIZ 6-carboxy-5 6 7 8-tetrahydropterin synthase OS=Candidatus Tokpelaia hoelldoblerii OX=1902579 GN=queD PE=3 SV=1
MM1 pKa = 7.42TYY3 pKa = 10.78VVTDD7 pKa = 2.96NCIRR11 pKa = 11.84CKK13 pKa = 10.23YY14 pKa = 7.51TDD16 pKa = 3.93CVEE19 pKa = 4.15VCPVDD24 pKa = 3.79CFYY27 pKa = 11.19EE28 pKa = 4.58GEE30 pKa = 4.21NMLVIHH36 pKa = 7.24PDD38 pKa = 2.97EE39 pKa = 5.88CIDD42 pKa = 4.44CGVCEE47 pKa = 4.44PEE49 pKa = 5.05CPAEE53 pKa = 4.42AIKK56 pKa = 10.65PDD58 pKa = 4.05TEE60 pKa = 4.72PGLDD64 pKa = 2.96QWLVLNAEE72 pKa = 4.8YY73 pKa = 10.26AAKK76 pKa = 9.48WPNITIKK83 pKa = 10.19KK84 pKa = 9.47DD85 pKa = 3.57QMPEE89 pKa = 3.57AAEE92 pKa = 3.95MDD94 pKa = 3.84GVPDD98 pKa = 3.19KK99 pKa = 10.99RR100 pKa = 11.84QYY102 pKa = 10.82FSEE105 pKa = 4.54KK106 pKa = 9.76PGSGDD111 pKa = 3.0
MM1 pKa = 7.42TYY3 pKa = 10.78VVTDD7 pKa = 2.96NCIRR11 pKa = 11.84CKK13 pKa = 10.23YY14 pKa = 7.51TDD16 pKa = 3.93CVEE19 pKa = 4.15VCPVDD24 pKa = 3.79CFYY27 pKa = 11.19EE28 pKa = 4.58GEE30 pKa = 4.21NMLVIHH36 pKa = 7.24PDD38 pKa = 2.97EE39 pKa = 5.88CIDD42 pKa = 4.44CGVCEE47 pKa = 4.44PEE49 pKa = 5.05CPAEE53 pKa = 4.42AIKK56 pKa = 10.65PDD58 pKa = 4.05TEE60 pKa = 4.72PGLDD64 pKa = 2.96QWLVLNAEE72 pKa = 4.8YY73 pKa = 10.26AAKK76 pKa = 9.48WPNITIKK83 pKa = 10.19KK84 pKa = 9.47DD85 pKa = 3.57QMPEE89 pKa = 3.57AAEE92 pKa = 3.95MDD94 pKa = 3.84GVPDD98 pKa = 3.19KK99 pKa = 10.99RR100 pKa = 11.84QYY102 pKa = 10.82FSEE105 pKa = 4.54KK106 pKa = 9.76PGSGDD111 pKa = 3.0
Molecular weight: 12.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U9JWG6|A0A1U9JWG6_9RHIZ DUF2293 domain-containing protein OS=Candidatus Tokpelaia hoelldoblerii OX=1902579 GN=BHV28_14900 PE=4 SV=1
MM1 pKa = 7.53RR2 pKa = 11.84GVRR5 pKa = 11.84RR6 pKa = 11.84IITKK10 pKa = 9.83AAAGGVQLMGMLLFPPGCPGCGRR33 pKa = 11.84AIAAIGTICSDD44 pKa = 3.51CWLEE48 pKa = 5.08LPFLTPPFCPVMGIPFHH65 pKa = 7.25SDD67 pKa = 2.87YY68 pKa = 11.63GDD70 pKa = 4.76GILSGEE76 pKa = 4.41ALASPPPFDD85 pKa = 3.32HH86 pKa = 6.75ARR88 pKa = 11.84AVVAHH93 pKa = 6.77EE94 pKa = 3.87GLARR98 pKa = 11.84NLVTRR103 pKa = 11.84LKK105 pKa = 10.71YY106 pKa = 10.19GARR109 pKa = 11.84TDD111 pKa = 3.85LAPWMATWMVRR122 pKa = 11.84AGYY125 pKa = 10.14DD126 pKa = 3.47LVSACDD132 pKa = 3.67MIVPVPLHH140 pKa = 6.53RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84FWKK146 pKa = 10.19RR147 pKa = 11.84GYY149 pKa = 9.39NQSAEE154 pKa = 4.21LARR157 pKa = 11.84AIARR161 pKa = 11.84ICGKK165 pKa = 8.96PFCPQLLARR174 pKa = 11.84RR175 pKa = 11.84KK176 pKa = 6.41PTRR179 pKa = 11.84PQVGLSAKK187 pKa = 9.79RR188 pKa = 11.84RR189 pKa = 11.84ADD191 pKa = 3.25NVRR194 pKa = 11.84GVFAVPPRR202 pKa = 11.84GAAALKK208 pKa = 9.78HH209 pKa = 4.82KK210 pKa = 10.39HH211 pKa = 5.69IVLVDD216 pKa = 3.45DD217 pKa = 4.81VYY219 pKa = 9.55TTGATVKK226 pKa = 10.31AAARR230 pKa = 11.84ILKK233 pKa = 9.83RR234 pKa = 11.84AGAARR239 pKa = 11.84VDD241 pKa = 3.51VLTFSRR247 pKa = 11.84VVIRR251 pKa = 11.84YY252 pKa = 7.25EE253 pKa = 3.67
MM1 pKa = 7.53RR2 pKa = 11.84GVRR5 pKa = 11.84RR6 pKa = 11.84IITKK10 pKa = 9.83AAAGGVQLMGMLLFPPGCPGCGRR33 pKa = 11.84AIAAIGTICSDD44 pKa = 3.51CWLEE48 pKa = 5.08LPFLTPPFCPVMGIPFHH65 pKa = 7.25SDD67 pKa = 2.87YY68 pKa = 11.63GDD70 pKa = 4.76GILSGEE76 pKa = 4.41ALASPPPFDD85 pKa = 3.32HH86 pKa = 6.75ARR88 pKa = 11.84AVVAHH93 pKa = 6.77EE94 pKa = 3.87GLARR98 pKa = 11.84NLVTRR103 pKa = 11.84LKK105 pKa = 10.71YY106 pKa = 10.19GARR109 pKa = 11.84TDD111 pKa = 3.85LAPWMATWMVRR122 pKa = 11.84AGYY125 pKa = 10.14DD126 pKa = 3.47LVSACDD132 pKa = 3.67MIVPVPLHH140 pKa = 6.53RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84FWKK146 pKa = 10.19RR147 pKa = 11.84GYY149 pKa = 9.39NQSAEE154 pKa = 4.21LARR157 pKa = 11.84AIARR161 pKa = 11.84ICGKK165 pKa = 8.96PFCPQLLARR174 pKa = 11.84RR175 pKa = 11.84KK176 pKa = 6.41PTRR179 pKa = 11.84PQVGLSAKK187 pKa = 9.79RR188 pKa = 11.84RR189 pKa = 11.84ADD191 pKa = 3.25NVRR194 pKa = 11.84GVFAVPPRR202 pKa = 11.84GAAALKK208 pKa = 9.78HH209 pKa = 4.82KK210 pKa = 10.39HH211 pKa = 5.69IVLVDD216 pKa = 3.45DD217 pKa = 4.81VYY219 pKa = 9.55TTGATVKK226 pKa = 10.31AAARR230 pKa = 11.84ILKK233 pKa = 9.83RR234 pKa = 11.84AGAARR239 pKa = 11.84VDD241 pKa = 3.51VLTFSRR247 pKa = 11.84VVIRR251 pKa = 11.84YY252 pKa = 7.25EE253 pKa = 3.67
Molecular weight: 27.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
556338 |
31 |
6177 |
330.6 |
36.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.415 ± 0.096 | 0.861 ± 0.021 |
5.461 ± 0.084 | 5.727 ± 0.064 |
3.986 ± 0.05 | 8.195 ± 0.089 |
2.159 ± 0.032 | 5.916 ± 0.05 |
4.347 ± 0.051 | 9.924 ± 0.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.655 ± 0.034 | 3.253 ± 0.058 |
4.469 ± 0.042 | 3.695 ± 0.038 |
6.446 ± 0.066 | 5.259 ± 0.046 |
5.31 ± 0.054 | 7.216 ± 0.051 |
1.164 ± 0.026 | 2.542 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
