
Paracoccus sp. J56
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus; unclassified Paracoccus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
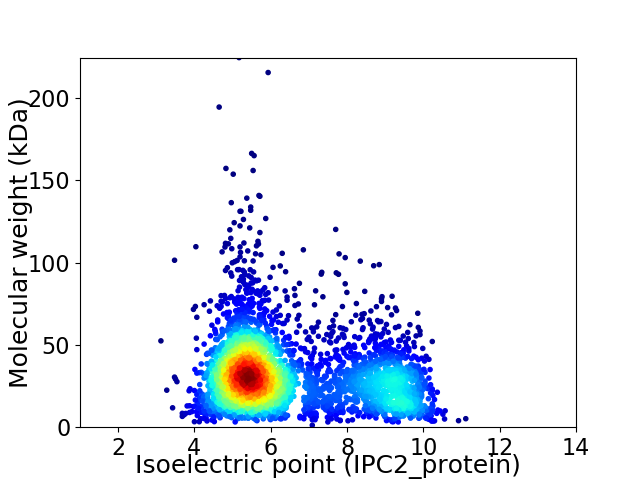
Virtual 2D-PAGE plot for 3752 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7IFA6|A0A1X7IFA6_9RHOB Mannose-binding protein /fructose-binding protein /ribose-binding protein OS=Paracoccus sp. J56 OX=935850 GN=SAMN02746000_00664 PE=4 SV=1
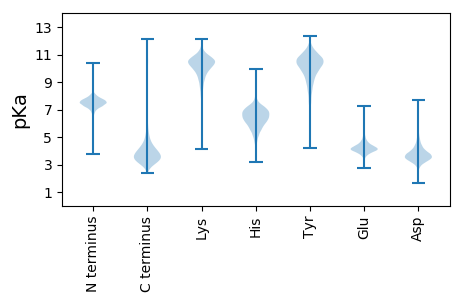
MM1 pKa = 7.18ATINGTDD8 pKa = 3.63GADD11 pKa = 3.56TLTGTPDD18 pKa = 3.42ADD20 pKa = 4.44VINGLAGNDD29 pKa = 3.86VITGGSGNDD38 pKa = 3.54TLRR41 pKa = 11.84GGSGDD46 pKa = 3.64DD47 pKa = 3.25TFLIGGTSPGTDD59 pKa = 2.66IFDD62 pKa = 4.29GGDD65 pKa = 3.23GADD68 pKa = 3.99RR69 pKa = 11.84IRR71 pKa = 11.84LNGNLTVSNLQLTSANVISTEE92 pKa = 3.92TLDD95 pKa = 3.57FSIYY99 pKa = 10.18SIQGTGANDD108 pKa = 3.43TFNISGITYY117 pKa = 8.34TSSYY121 pKa = 11.14DD122 pKa = 2.87WFYY125 pKa = 11.48LYY127 pKa = 10.68DD128 pKa = 4.24GNDD131 pKa = 3.15SFVGYY136 pKa = 10.02VGNDD140 pKa = 3.4SVDD143 pKa = 3.24GGAGNDD149 pKa = 3.93TLNGGAGNDD158 pKa = 3.54VLTGGGGNDD167 pKa = 3.61HH168 pKa = 7.36LIGGSGNDD176 pKa = 3.29TFQIGGTDD184 pKa = 2.8IGYY187 pKa = 9.82DD188 pKa = 3.2IYY190 pKa = 11.62NGGDD194 pKa = 3.07GDD196 pKa = 4.16DD197 pKa = 4.47RR198 pKa = 11.84IRR200 pKa = 11.84LTSNVTVSNLQLTSANVISTEE221 pKa = 3.92TLDD224 pKa = 3.57FSIYY228 pKa = 10.18SIQGTGANDD237 pKa = 3.43TFNISGITYY246 pKa = 8.34TSSYY250 pKa = 11.14DD251 pKa = 2.87WFYY254 pKa = 11.48LYY256 pKa = 10.68DD257 pKa = 4.24GNDD260 pKa = 3.15SFVGYY265 pKa = 10.02VGNDD269 pKa = 3.4SVDD272 pKa = 3.24GGAGNDD278 pKa = 3.93TLNGGAGNDD287 pKa = 3.54VLTGGGGNDD296 pKa = 3.61HH297 pKa = 7.36LIGGSGNDD305 pKa = 3.29TFQIGGTDD313 pKa = 2.8IGYY316 pKa = 9.82DD317 pKa = 3.2IYY319 pKa = 11.62NGGDD323 pKa = 3.07GDD325 pKa = 4.16DD326 pKa = 4.47RR327 pKa = 11.84IRR329 pKa = 11.84LTSNVTVSNLQLTSANVISTEE350 pKa = 3.92TLDD353 pKa = 3.57FSIYY357 pKa = 10.18SIQGTGANDD366 pKa = 3.43TFNISGITYY375 pKa = 7.83TSSYY379 pKa = 11.0DD380 pKa = 3.05RR381 pKa = 11.84FYY383 pKa = 10.88MHH385 pKa = 7.78DD386 pKa = 3.96GNDD389 pKa = 3.26SFIGYY394 pKa = 9.19SGNDD398 pKa = 3.42AVDD401 pKa = 3.79GGAGNDD407 pKa = 3.78TLRR410 pKa = 11.84GEE412 pKa = 4.57AGNDD416 pKa = 3.28QLIGGSGNDD425 pKa = 3.41VLIGGTGNDD434 pKa = 3.84TLDD437 pKa = 3.87GGTGSDD443 pKa = 3.0TALYY447 pKa = 10.57DD448 pKa = 3.65GATSGVRR455 pKa = 11.84VNLGSTSAQAIGGGQGTDD473 pKa = 2.79VLTSIEE479 pKa = 4.16HH480 pKa = 5.18VRR482 pKa = 11.84GSRR485 pKa = 11.84YY486 pKa = 9.75SDD488 pKa = 3.29SLTGSGASNVLNAGAGNDD506 pKa = 4.03TLSGGAGNDD515 pKa = 3.29RR516 pKa = 11.84LIGGDD521 pKa = 3.4GNDD524 pKa = 3.65YY525 pKa = 11.22LKK527 pKa = 11.18GGTGNDD533 pKa = 3.63TLEE536 pKa = 4.69GGAGTDD542 pKa = 3.16TALYY546 pKa = 10.47DD547 pKa = 3.69SATGGVKK554 pKa = 10.42VNLGSASAQAIGGGQGTDD572 pKa = 2.86VLTGIEE578 pKa = 4.08NVRR581 pKa = 11.84GSRR584 pKa = 11.84YY585 pKa = 9.73NDD587 pKa = 3.24SLTGSGGNNILNAGAGNDD605 pKa = 4.03TLSGGAGNDD614 pKa = 3.39RR615 pKa = 11.84LIGGAGNDD623 pKa = 3.78YY624 pKa = 11.27LNGGTGNDD632 pKa = 3.79TLDD635 pKa = 3.58GSTGSDD641 pKa = 2.72TVLYY645 pKa = 10.6DD646 pKa = 3.45GATSAVTVNLGSTKK660 pKa = 10.37AQAIGGGYY668 pKa = 8.17GTDD671 pKa = 3.02VLSSIEE677 pKa = 3.93NVRR680 pKa = 11.84GSRR683 pKa = 11.84YY684 pKa = 9.67SDD686 pKa = 3.32SLTGSGGNNVLNAGAGNDD704 pKa = 4.03TLSGGAGSDD713 pKa = 3.47RR714 pKa = 11.84LIGGSGNDD722 pKa = 3.62YY723 pKa = 11.24LNGGTGNDD731 pKa = 3.62TLEE734 pKa = 4.85GGTGTDD740 pKa = 2.94TVLYY744 pKa = 10.38DD745 pKa = 3.61GATSAVTINLGSTKK759 pKa = 10.27AQAIGGGYY767 pKa = 8.17GTDD770 pKa = 3.02VLSSIEE776 pKa = 3.93NVRR779 pKa = 11.84GSRR782 pKa = 11.84YY783 pKa = 9.67SDD785 pKa = 3.32SLTGSGGNNVLNAGAGNDD803 pKa = 4.03TLSGGAGNDD812 pKa = 3.32RR813 pKa = 11.84LIGGSGNDD821 pKa = 3.62YY822 pKa = 11.24LNGGTGNDD830 pKa = 3.77TLDD833 pKa = 3.73GGTGIDD839 pKa = 3.17TALYY843 pKa = 10.57DD844 pKa = 4.03GATSGIKK851 pKa = 10.38VNLGSTSAQAIGGGYY866 pKa = 8.08GTDD869 pKa = 3.01VLNSIEE875 pKa = 4.26NIRR878 pKa = 11.84GSRR881 pKa = 11.84YY882 pKa = 9.57SDD884 pKa = 3.3SLTGSGANNVLNAGAGNDD902 pKa = 3.99TVSGGAGNDD911 pKa = 3.36QLIGGTGNDD920 pKa = 3.47RR921 pKa = 11.84LLGGTGNDD929 pKa = 3.77TLTGGAGNDD938 pKa = 3.62VLSGGSGADD947 pKa = 2.85VFVFSVRR954 pKa = 11.84DD955 pKa = 3.48NADD958 pKa = 4.85RR959 pKa = 11.84ISDD962 pKa = 3.67WQNGSDD968 pKa = 4.15VIRR971 pKa = 11.84IVAGTYY977 pKa = 10.08DD978 pKa = 3.53GEE980 pKa = 4.78RR981 pKa = 11.84YY982 pKa = 10.4DD983 pKa = 5.18SFDD986 pKa = 4.77DD987 pKa = 3.97LTITQSGSNTVVSFGGTTVTLTGINSGLLDD1017 pKa = 3.81ASDD1020 pKa = 3.93FVFVV1024 pKa = 4.16
MM1 pKa = 7.18ATINGTDD8 pKa = 3.63GADD11 pKa = 3.56TLTGTPDD18 pKa = 3.42ADD20 pKa = 4.44VINGLAGNDD29 pKa = 3.86VITGGSGNDD38 pKa = 3.54TLRR41 pKa = 11.84GGSGDD46 pKa = 3.64DD47 pKa = 3.25TFLIGGTSPGTDD59 pKa = 2.66IFDD62 pKa = 4.29GGDD65 pKa = 3.23GADD68 pKa = 3.99RR69 pKa = 11.84IRR71 pKa = 11.84LNGNLTVSNLQLTSANVISTEE92 pKa = 3.92TLDD95 pKa = 3.57FSIYY99 pKa = 10.18SIQGTGANDD108 pKa = 3.43TFNISGITYY117 pKa = 8.34TSSYY121 pKa = 11.14DD122 pKa = 2.87WFYY125 pKa = 11.48LYY127 pKa = 10.68DD128 pKa = 4.24GNDD131 pKa = 3.15SFVGYY136 pKa = 10.02VGNDD140 pKa = 3.4SVDD143 pKa = 3.24GGAGNDD149 pKa = 3.93TLNGGAGNDD158 pKa = 3.54VLTGGGGNDD167 pKa = 3.61HH168 pKa = 7.36LIGGSGNDD176 pKa = 3.29TFQIGGTDD184 pKa = 2.8IGYY187 pKa = 9.82DD188 pKa = 3.2IYY190 pKa = 11.62NGGDD194 pKa = 3.07GDD196 pKa = 4.16DD197 pKa = 4.47RR198 pKa = 11.84IRR200 pKa = 11.84LTSNVTVSNLQLTSANVISTEE221 pKa = 3.92TLDD224 pKa = 3.57FSIYY228 pKa = 10.18SIQGTGANDD237 pKa = 3.43TFNISGITYY246 pKa = 8.34TSSYY250 pKa = 11.14DD251 pKa = 2.87WFYY254 pKa = 11.48LYY256 pKa = 10.68DD257 pKa = 4.24GNDD260 pKa = 3.15SFVGYY265 pKa = 10.02VGNDD269 pKa = 3.4SVDD272 pKa = 3.24GGAGNDD278 pKa = 3.93TLNGGAGNDD287 pKa = 3.54VLTGGGGNDD296 pKa = 3.61HH297 pKa = 7.36LIGGSGNDD305 pKa = 3.29TFQIGGTDD313 pKa = 2.8IGYY316 pKa = 9.82DD317 pKa = 3.2IYY319 pKa = 11.62NGGDD323 pKa = 3.07GDD325 pKa = 4.16DD326 pKa = 4.47RR327 pKa = 11.84IRR329 pKa = 11.84LTSNVTVSNLQLTSANVISTEE350 pKa = 3.92TLDD353 pKa = 3.57FSIYY357 pKa = 10.18SIQGTGANDD366 pKa = 3.43TFNISGITYY375 pKa = 7.83TSSYY379 pKa = 11.0DD380 pKa = 3.05RR381 pKa = 11.84FYY383 pKa = 10.88MHH385 pKa = 7.78DD386 pKa = 3.96GNDD389 pKa = 3.26SFIGYY394 pKa = 9.19SGNDD398 pKa = 3.42AVDD401 pKa = 3.79GGAGNDD407 pKa = 3.78TLRR410 pKa = 11.84GEE412 pKa = 4.57AGNDD416 pKa = 3.28QLIGGSGNDD425 pKa = 3.41VLIGGTGNDD434 pKa = 3.84TLDD437 pKa = 3.87GGTGSDD443 pKa = 3.0TALYY447 pKa = 10.57DD448 pKa = 3.65GATSGVRR455 pKa = 11.84VNLGSTSAQAIGGGQGTDD473 pKa = 2.79VLTSIEE479 pKa = 4.16HH480 pKa = 5.18VRR482 pKa = 11.84GSRR485 pKa = 11.84YY486 pKa = 9.75SDD488 pKa = 3.29SLTGSGASNVLNAGAGNDD506 pKa = 4.03TLSGGAGNDD515 pKa = 3.29RR516 pKa = 11.84LIGGDD521 pKa = 3.4GNDD524 pKa = 3.65YY525 pKa = 11.22LKK527 pKa = 11.18GGTGNDD533 pKa = 3.63TLEE536 pKa = 4.69GGAGTDD542 pKa = 3.16TALYY546 pKa = 10.47DD547 pKa = 3.69SATGGVKK554 pKa = 10.42VNLGSASAQAIGGGQGTDD572 pKa = 2.86VLTGIEE578 pKa = 4.08NVRR581 pKa = 11.84GSRR584 pKa = 11.84YY585 pKa = 9.73NDD587 pKa = 3.24SLTGSGGNNILNAGAGNDD605 pKa = 4.03TLSGGAGNDD614 pKa = 3.39RR615 pKa = 11.84LIGGAGNDD623 pKa = 3.78YY624 pKa = 11.27LNGGTGNDD632 pKa = 3.79TLDD635 pKa = 3.58GSTGSDD641 pKa = 2.72TVLYY645 pKa = 10.6DD646 pKa = 3.45GATSAVTVNLGSTKK660 pKa = 10.37AQAIGGGYY668 pKa = 8.17GTDD671 pKa = 3.02VLSSIEE677 pKa = 3.93NVRR680 pKa = 11.84GSRR683 pKa = 11.84YY684 pKa = 9.67SDD686 pKa = 3.32SLTGSGGNNVLNAGAGNDD704 pKa = 4.03TLSGGAGSDD713 pKa = 3.47RR714 pKa = 11.84LIGGSGNDD722 pKa = 3.62YY723 pKa = 11.24LNGGTGNDD731 pKa = 3.62TLEE734 pKa = 4.85GGTGTDD740 pKa = 2.94TVLYY744 pKa = 10.38DD745 pKa = 3.61GATSAVTINLGSTKK759 pKa = 10.27AQAIGGGYY767 pKa = 8.17GTDD770 pKa = 3.02VLSSIEE776 pKa = 3.93NVRR779 pKa = 11.84GSRR782 pKa = 11.84YY783 pKa = 9.67SDD785 pKa = 3.32SLTGSGGNNVLNAGAGNDD803 pKa = 4.03TLSGGAGNDD812 pKa = 3.32RR813 pKa = 11.84LIGGSGNDD821 pKa = 3.62YY822 pKa = 11.24LNGGTGNDD830 pKa = 3.77TLDD833 pKa = 3.73GGTGIDD839 pKa = 3.17TALYY843 pKa = 10.57DD844 pKa = 4.03GATSGIKK851 pKa = 10.38VNLGSTSAQAIGGGYY866 pKa = 8.08GTDD869 pKa = 3.01VLNSIEE875 pKa = 4.26NIRR878 pKa = 11.84GSRR881 pKa = 11.84YY882 pKa = 9.57SDD884 pKa = 3.3SLTGSGANNVLNAGAGNDD902 pKa = 3.99TVSGGAGNDD911 pKa = 3.36QLIGGTGNDD920 pKa = 3.47RR921 pKa = 11.84LLGGTGNDD929 pKa = 3.77TLTGGAGNDD938 pKa = 3.62VLSGGSGADD947 pKa = 2.85VFVFSVRR954 pKa = 11.84DD955 pKa = 3.48NADD958 pKa = 4.85RR959 pKa = 11.84ISDD962 pKa = 3.67WQNGSDD968 pKa = 4.15VIRR971 pKa = 11.84IVAGTYY977 pKa = 10.08DD978 pKa = 3.53GEE980 pKa = 4.78RR981 pKa = 11.84YY982 pKa = 10.4DD983 pKa = 5.18SFDD986 pKa = 4.77DD987 pKa = 3.97LTITQSGSNTVVSFGGTTVTLTGINSGLLDD1017 pKa = 3.81ASDD1020 pKa = 3.93FVFVV1024 pKa = 4.16
Molecular weight: 101.44 kDa
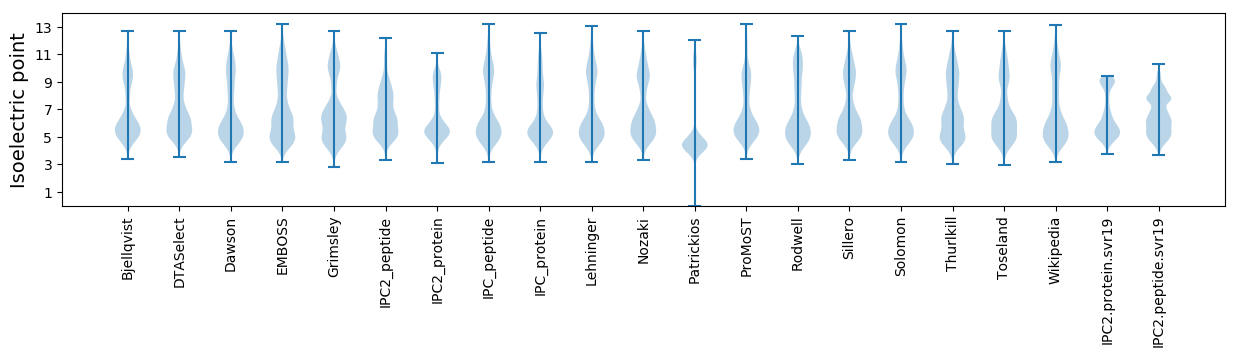
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7KEG8|A0A1X7KEG8_9RHOB Putative spermidine/putrescine transport system permease protein OS=Paracoccus sp. J56 OX=935850 GN=SAMN02746000_02353 PE=3 SV=1
MM1 pKa = 7.0TLIWMTAIAMVSAGMVALRR20 pKa = 11.84LRR22 pKa = 11.84APQPQAVAARR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 3.41
MM1 pKa = 7.0TLIWMTAIAMVSAGMVALRR20 pKa = 11.84LRR22 pKa = 11.84APQPQAVAARR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 3.41
Molecular weight: 3.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1148692 |
11 |
2099 |
306.2 |
33.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.762 ± 0.059 | 0.862 ± 0.012 |
5.606 ± 0.03 | 5.845 ± 0.038 |
3.481 ± 0.026 | 8.888 ± 0.043 |
2.098 ± 0.019 | 5.267 ± 0.028 |
2.699 ± 0.029 | 10.421 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.698 ± 0.02 | 2.408 ± 0.019 |
5.489 ± 0.03 | 3.295 ± 0.021 |
7.584 ± 0.042 | 4.954 ± 0.026 |
5.117 ± 0.026 | 7.047 ± 0.035 |
1.439 ± 0.015 | 2.041 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
