
Sphingomonas sp. YR710
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
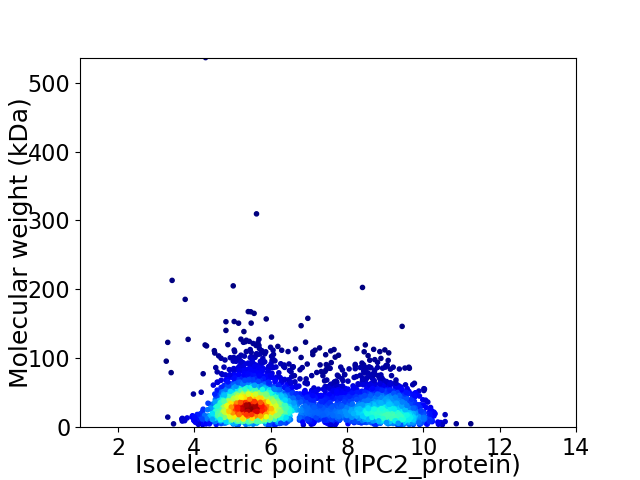
Virtual 2D-PAGE plot for 4037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6V8A7|A0A1G6V8A7_9SPHN Uncharacterized protein OS=Sphingomonas sp. YR710 OX=1882773 GN=SAMN05444678_11463 PE=4 SV=1
MM1 pKa = 7.12ATSAPVIYY9 pKa = 10.72DD10 pKa = 3.32GDD12 pKa = 3.89LSDD15 pKa = 3.68WSDD18 pKa = 3.71SEE20 pKa = 5.63RR21 pKa = 11.84IDD23 pKa = 3.99NAIEE27 pKa = 4.19SPPGYY32 pKa = 10.53ALYY35 pKa = 10.59ARR37 pKa = 11.84PDD39 pKa = 3.54GSSLAFAFHH48 pKa = 6.33SAVPIGANTTFWLNTDD64 pKa = 3.41RR65 pKa = 11.84DD66 pKa = 3.94ATTGYY71 pKa = 10.59QIFGFAGGAEE81 pKa = 4.27YY82 pKa = 10.78NVNIAADD89 pKa = 4.29GTASLYY95 pKa = 10.34TGAAGQTLVLANLQIAYY112 pKa = 10.04NADD115 pKa = 3.12HH116 pKa = 6.37TVAEE120 pKa = 4.75FSLPKK125 pKa = 10.17AAIGNPPAIDD135 pKa = 3.27VLYY138 pKa = 10.51DD139 pKa = 3.29VNDD142 pKa = 3.63AVFGPTSYY150 pKa = 11.0SDD152 pKa = 3.75KK153 pKa = 10.95PFTLFNDD160 pKa = 3.55TGIAPNLSTKK170 pKa = 10.57VGIVYY175 pKa = 10.34SDD177 pKa = 3.55TTAHH181 pKa = 6.18QYY183 pKa = 10.55FSEE186 pKa = 4.17TAYY189 pKa = 10.77AQLFMNAQAQAMQAGVSFDD208 pKa = 4.31LLSEE212 pKa = 4.29SDD214 pKa = 3.76LTDD217 pKa = 3.69LSKK220 pKa = 10.66ISQYY224 pKa = 10.03DD225 pKa = 3.46TLIFPSFRR233 pKa = 11.84NVQASQANAIANTLQQAEE251 pKa = 4.43VQFGVGLIVGGEE263 pKa = 4.0FMTDD267 pKa = 3.4DD268 pKa = 4.37EE269 pKa = 5.22NNNPLPGDD277 pKa = 3.6SYY279 pKa = 12.15SRR281 pKa = 11.84MKK283 pKa = 10.92LLLDD287 pKa = 3.48ATRR290 pKa = 11.84VTGGTGDD297 pKa = 3.65VSLQATDD304 pKa = 3.44PGGLVLQNYY313 pKa = 9.28ASNEE317 pKa = 3.96VIRR320 pKa = 11.84NYY322 pKa = 10.53TGVGWNAFTNVSGTGTTVATEE343 pKa = 4.44TIGGQTYY350 pKa = 9.43AAAMATQTGGRR361 pKa = 11.84NVVFSSEE368 pKa = 3.48ATMADD373 pKa = 5.18SNLLWQAIDD382 pKa = 3.78YY383 pKa = 8.47SAQGTGVSVGLQLTRR398 pKa = 11.84QTGIVAARR406 pKa = 11.84VDD408 pKa = 4.91LDD410 pKa = 3.34SSMYY414 pKa = 11.07AEE416 pKa = 5.3DD417 pKa = 4.19VNPLDD422 pKa = 4.54GSPGVYY428 pKa = 10.13DD429 pKa = 3.56KK430 pKa = 11.23LVPILAQWKK439 pKa = 9.21DD440 pKa = 2.98AYY442 pKa = 11.22NFVGSLYY449 pKa = 11.15ANIGNNPAQGIYY461 pKa = 10.12TDD463 pKa = 3.59WTVSLPYY470 pKa = 9.43YY471 pKa = 10.46KK472 pKa = 11.21AMIDD476 pKa = 3.57LGNEE480 pKa = 3.99IGTHH484 pKa = 6.22SYY486 pKa = 7.96THH488 pKa = 7.45PDD490 pKa = 3.09NTNLLSAAQIQTEE503 pKa = 4.45FADD506 pKa = 3.4STTILNQQLSAYY518 pKa = 9.73LGHH521 pKa = 7.74PYY523 pKa = 10.34NIVGAAVPGAPEE535 pKa = 3.88TVATSTEE542 pKa = 3.38IMKK545 pKa = 10.09YY546 pKa = 7.58VQQYY550 pKa = 7.98LTGGYY555 pKa = 9.14SGVGAGYY562 pKa = 9.45PDD564 pKa = 3.57AFGFLSPTQEE574 pKa = 3.86NKK576 pKa = 10.22VYY578 pKa = 10.45LAPNTSFDD586 pKa = 3.47FTLVEE591 pKa = 4.45FQGKK595 pKa = 8.13TIAQAEE601 pKa = 4.32AAWAQEE607 pKa = 3.98WTDD610 pKa = 3.9LTSKK614 pKa = 10.42GQTPIVVWPFHH625 pKa = 7.41DD626 pKa = 3.82YY627 pKa = 10.6AAAAFDD633 pKa = 4.02VNGTGTSPYY642 pKa = 6.39TTQMFTDD649 pKa = 4.86WIARR653 pKa = 11.84ASASGSEE660 pKa = 4.27FVTLADD666 pKa = 3.82LADD669 pKa = 5.6RR670 pKa = 11.84IQAFDD675 pKa = 3.51QASVTTSVVGNTITATVGSSDD696 pKa = 3.4AGRR699 pKa = 11.84FALDD703 pKa = 3.6LQRR706 pKa = 11.84QGSSVIQNVDD716 pKa = 1.94NWYY719 pKa = 10.47AYY721 pKa = 8.58DD722 pKa = 3.65TDD724 pKa = 5.01SIFMPASGGTFTIHH738 pKa = 7.03LGAAQDD744 pKa = 4.12DD745 pKa = 4.44VTHH748 pKa = 6.8IAALPMRR755 pKa = 11.84ASLISVAGDD764 pKa = 3.25GRR766 pKa = 11.84NLAFSVEE773 pKa = 3.97GDD775 pKa = 3.63GLVTVDD781 pKa = 4.42LQNPGTDD788 pKa = 4.38LINVTGATIASQSGEE803 pKa = 4.03ILTLNLGAAGHH814 pKa = 6.69HH815 pKa = 6.94DD816 pKa = 3.65VTIGLGADD824 pKa = 3.49SAPVITSNGGGDD836 pKa = 3.51TAALSIPEE844 pKa = 4.73HH845 pKa = 6.07IAAVATVVATDD856 pKa = 4.08ANAGDD861 pKa = 3.29ITTYY865 pKa = 10.93GIASGVDD872 pKa = 3.08ASLFAIDD879 pKa = 4.38SVTGVLSFKK888 pKa = 10.35VAPDD892 pKa = 3.22YY893 pKa = 9.52TLPQDD898 pKa = 3.63SDD900 pKa = 3.97HH901 pKa = 6.55NNSYY905 pKa = 10.51LVNVSATDD913 pKa = 3.38SHH915 pKa = 6.93GLADD919 pKa = 4.45LQQLTVNVTNANVAPIITSNGAGATATLGIAEE951 pKa = 4.46NTTAVTMVVATDD963 pKa = 3.62ANRR966 pKa = 11.84ATGDD970 pKa = 3.29TTTYY974 pKa = 10.62GIVSGADD981 pKa = 2.98AALFAIDD988 pKa = 4.47PVTGVLTFKK997 pKa = 10.7AAPDD1001 pKa = 3.59YY1002 pKa = 9.98EE1003 pKa = 4.44LPQDD1007 pKa = 3.66SDD1009 pKa = 3.96HH1010 pKa = 6.57NNSYY1014 pKa = 10.6LVNISATDD1022 pKa = 3.47SRR1024 pKa = 11.84GLADD1028 pKa = 4.37LQNLTVNVTNVLGIVQTGTSGNDD1051 pKa = 2.5IMTGTIEE1058 pKa = 4.83PDD1060 pKa = 3.44TLSGAGGADD1069 pKa = 3.77TISGLGGNDD1078 pKa = 2.89ILNGNAGKK1086 pKa = 10.21DD1087 pKa = 3.5IINGGVGNDD1096 pKa = 4.2RR1097 pKa = 11.84ITGGAGADD1105 pKa = 3.75TLTGGPGNDD1114 pKa = 2.93LFVYY1118 pKa = 10.28TDD1120 pKa = 3.44VADD1123 pKa = 4.05SGTTATTRR1131 pKa = 11.84DD1132 pKa = 3.55VITDD1136 pKa = 3.96FEE1138 pKa = 4.38HH1139 pKa = 7.45AIDD1142 pKa = 5.68RR1143 pKa = 11.84IDD1145 pKa = 4.98LSAIDD1150 pKa = 4.15ANAGRR1155 pKa = 11.84GSLRR1159 pKa = 11.84GDD1161 pKa = 3.22QAFTFLPVAGTAFTGAAQLRR1181 pKa = 11.84YY1182 pKa = 8.29TYY1184 pKa = 9.26VTVGGVEE1191 pKa = 3.91HH1192 pKa = 6.91TIVEE1196 pKa = 4.09GDD1198 pKa = 3.66VNVNHH1203 pKa = 7.07IGADD1207 pKa = 3.67FSIDD1211 pKa = 3.68LVGHH1215 pKa = 6.27QILTASDD1222 pKa = 4.95FILL1225 pKa = 4.96
MM1 pKa = 7.12ATSAPVIYY9 pKa = 10.72DD10 pKa = 3.32GDD12 pKa = 3.89LSDD15 pKa = 3.68WSDD18 pKa = 3.71SEE20 pKa = 5.63RR21 pKa = 11.84IDD23 pKa = 3.99NAIEE27 pKa = 4.19SPPGYY32 pKa = 10.53ALYY35 pKa = 10.59ARR37 pKa = 11.84PDD39 pKa = 3.54GSSLAFAFHH48 pKa = 6.33SAVPIGANTTFWLNTDD64 pKa = 3.41RR65 pKa = 11.84DD66 pKa = 3.94ATTGYY71 pKa = 10.59QIFGFAGGAEE81 pKa = 4.27YY82 pKa = 10.78NVNIAADD89 pKa = 4.29GTASLYY95 pKa = 10.34TGAAGQTLVLANLQIAYY112 pKa = 10.04NADD115 pKa = 3.12HH116 pKa = 6.37TVAEE120 pKa = 4.75FSLPKK125 pKa = 10.17AAIGNPPAIDD135 pKa = 3.27VLYY138 pKa = 10.51DD139 pKa = 3.29VNDD142 pKa = 3.63AVFGPTSYY150 pKa = 11.0SDD152 pKa = 3.75KK153 pKa = 10.95PFTLFNDD160 pKa = 3.55TGIAPNLSTKK170 pKa = 10.57VGIVYY175 pKa = 10.34SDD177 pKa = 3.55TTAHH181 pKa = 6.18QYY183 pKa = 10.55FSEE186 pKa = 4.17TAYY189 pKa = 10.77AQLFMNAQAQAMQAGVSFDD208 pKa = 4.31LLSEE212 pKa = 4.29SDD214 pKa = 3.76LTDD217 pKa = 3.69LSKK220 pKa = 10.66ISQYY224 pKa = 10.03DD225 pKa = 3.46TLIFPSFRR233 pKa = 11.84NVQASQANAIANTLQQAEE251 pKa = 4.43VQFGVGLIVGGEE263 pKa = 4.0FMTDD267 pKa = 3.4DD268 pKa = 4.37EE269 pKa = 5.22NNNPLPGDD277 pKa = 3.6SYY279 pKa = 12.15SRR281 pKa = 11.84MKK283 pKa = 10.92LLLDD287 pKa = 3.48ATRR290 pKa = 11.84VTGGTGDD297 pKa = 3.65VSLQATDD304 pKa = 3.44PGGLVLQNYY313 pKa = 9.28ASNEE317 pKa = 3.96VIRR320 pKa = 11.84NYY322 pKa = 10.53TGVGWNAFTNVSGTGTTVATEE343 pKa = 4.44TIGGQTYY350 pKa = 9.43AAAMATQTGGRR361 pKa = 11.84NVVFSSEE368 pKa = 3.48ATMADD373 pKa = 5.18SNLLWQAIDD382 pKa = 3.78YY383 pKa = 8.47SAQGTGVSVGLQLTRR398 pKa = 11.84QTGIVAARR406 pKa = 11.84VDD408 pKa = 4.91LDD410 pKa = 3.34SSMYY414 pKa = 11.07AEE416 pKa = 5.3DD417 pKa = 4.19VNPLDD422 pKa = 4.54GSPGVYY428 pKa = 10.13DD429 pKa = 3.56KK430 pKa = 11.23LVPILAQWKK439 pKa = 9.21DD440 pKa = 2.98AYY442 pKa = 11.22NFVGSLYY449 pKa = 11.15ANIGNNPAQGIYY461 pKa = 10.12TDD463 pKa = 3.59WTVSLPYY470 pKa = 9.43YY471 pKa = 10.46KK472 pKa = 11.21AMIDD476 pKa = 3.57LGNEE480 pKa = 3.99IGTHH484 pKa = 6.22SYY486 pKa = 7.96THH488 pKa = 7.45PDD490 pKa = 3.09NTNLLSAAQIQTEE503 pKa = 4.45FADD506 pKa = 3.4STTILNQQLSAYY518 pKa = 9.73LGHH521 pKa = 7.74PYY523 pKa = 10.34NIVGAAVPGAPEE535 pKa = 3.88TVATSTEE542 pKa = 3.38IMKK545 pKa = 10.09YY546 pKa = 7.58VQQYY550 pKa = 7.98LTGGYY555 pKa = 9.14SGVGAGYY562 pKa = 9.45PDD564 pKa = 3.57AFGFLSPTQEE574 pKa = 3.86NKK576 pKa = 10.22VYY578 pKa = 10.45LAPNTSFDD586 pKa = 3.47FTLVEE591 pKa = 4.45FQGKK595 pKa = 8.13TIAQAEE601 pKa = 4.32AAWAQEE607 pKa = 3.98WTDD610 pKa = 3.9LTSKK614 pKa = 10.42GQTPIVVWPFHH625 pKa = 7.41DD626 pKa = 3.82YY627 pKa = 10.6AAAAFDD633 pKa = 4.02VNGTGTSPYY642 pKa = 6.39TTQMFTDD649 pKa = 4.86WIARR653 pKa = 11.84ASASGSEE660 pKa = 4.27FVTLADD666 pKa = 3.82LADD669 pKa = 5.6RR670 pKa = 11.84IQAFDD675 pKa = 3.51QASVTTSVVGNTITATVGSSDD696 pKa = 3.4AGRR699 pKa = 11.84FALDD703 pKa = 3.6LQRR706 pKa = 11.84QGSSVIQNVDD716 pKa = 1.94NWYY719 pKa = 10.47AYY721 pKa = 8.58DD722 pKa = 3.65TDD724 pKa = 5.01SIFMPASGGTFTIHH738 pKa = 7.03LGAAQDD744 pKa = 4.12DD745 pKa = 4.44VTHH748 pKa = 6.8IAALPMRR755 pKa = 11.84ASLISVAGDD764 pKa = 3.25GRR766 pKa = 11.84NLAFSVEE773 pKa = 3.97GDD775 pKa = 3.63GLVTVDD781 pKa = 4.42LQNPGTDD788 pKa = 4.38LINVTGATIASQSGEE803 pKa = 4.03ILTLNLGAAGHH814 pKa = 6.69HH815 pKa = 6.94DD816 pKa = 3.65VTIGLGADD824 pKa = 3.49SAPVITSNGGGDD836 pKa = 3.51TAALSIPEE844 pKa = 4.73HH845 pKa = 6.07IAAVATVVATDD856 pKa = 4.08ANAGDD861 pKa = 3.29ITTYY865 pKa = 10.93GIASGVDD872 pKa = 3.08ASLFAIDD879 pKa = 4.38SVTGVLSFKK888 pKa = 10.35VAPDD892 pKa = 3.22YY893 pKa = 9.52TLPQDD898 pKa = 3.63SDD900 pKa = 3.97HH901 pKa = 6.55NNSYY905 pKa = 10.51LVNVSATDD913 pKa = 3.38SHH915 pKa = 6.93GLADD919 pKa = 4.45LQQLTVNVTNANVAPIITSNGAGATATLGIAEE951 pKa = 4.46NTTAVTMVVATDD963 pKa = 3.62ANRR966 pKa = 11.84ATGDD970 pKa = 3.29TTTYY974 pKa = 10.62GIVSGADD981 pKa = 2.98AALFAIDD988 pKa = 4.47PVTGVLTFKK997 pKa = 10.7AAPDD1001 pKa = 3.59YY1002 pKa = 9.98EE1003 pKa = 4.44LPQDD1007 pKa = 3.66SDD1009 pKa = 3.96HH1010 pKa = 6.57NNSYY1014 pKa = 10.6LVNISATDD1022 pKa = 3.47SRR1024 pKa = 11.84GLADD1028 pKa = 4.37LQNLTVNVTNVLGIVQTGTSGNDD1051 pKa = 2.5IMTGTIEE1058 pKa = 4.83PDD1060 pKa = 3.44TLSGAGGADD1069 pKa = 3.77TISGLGGNDD1078 pKa = 2.89ILNGNAGKK1086 pKa = 10.21DD1087 pKa = 3.5IINGGVGNDD1096 pKa = 4.2RR1097 pKa = 11.84ITGGAGADD1105 pKa = 3.75TLTGGPGNDD1114 pKa = 2.93LFVYY1118 pKa = 10.28TDD1120 pKa = 3.44VADD1123 pKa = 4.05SGTTATTRR1131 pKa = 11.84DD1132 pKa = 3.55VITDD1136 pKa = 3.96FEE1138 pKa = 4.38HH1139 pKa = 7.45AIDD1142 pKa = 5.68RR1143 pKa = 11.84IDD1145 pKa = 4.98LSAIDD1150 pKa = 4.15ANAGRR1155 pKa = 11.84GSLRR1159 pKa = 11.84GDD1161 pKa = 3.22QAFTFLPVAGTAFTGAAQLRR1181 pKa = 11.84YY1182 pKa = 8.29TYY1184 pKa = 9.26VTVGGVEE1191 pKa = 3.91HH1192 pKa = 6.91TIVEE1196 pKa = 4.09GDD1198 pKa = 3.66VNVNHH1203 pKa = 7.07IGADD1207 pKa = 3.67FSIDD1211 pKa = 3.68LVGHH1215 pKa = 6.27QILTASDD1222 pKa = 4.95FILL1225 pKa = 4.96
Molecular weight: 127.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6I3Z6|A0A1G6I3Z6_9SPHN Uncharacterized conserved protein DUF885 familyt OS=Sphingomonas sp. YR710 OX=1882773 GN=SAMN05444678_101111 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.53GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.45LSAA44 pKa = 3.94
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.53GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.45LSAA44 pKa = 3.94
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1304086 |
28 |
5710 |
323.0 |
34.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.313 ± 0.06 | 0.791 ± 0.011 |
6.026 ± 0.035 | 4.936 ± 0.042 |
3.539 ± 0.026 | 8.819 ± 0.049 |
2.087 ± 0.021 | 5.696 ± 0.025 |
2.913 ± 0.03 | 9.884 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.022 | 2.554 ± 0.03 |
5.251 ± 0.034 | 2.994 ± 0.022 |
7.278 ± 0.046 | 5.499 ± 0.052 |
5.341 ± 0.053 | 7.007 ± 0.029 |
1.333 ± 0.02 | 2.208 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
