
Leifsonia sp. Root112D2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Leifsonia; unclassified Leifsonia
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
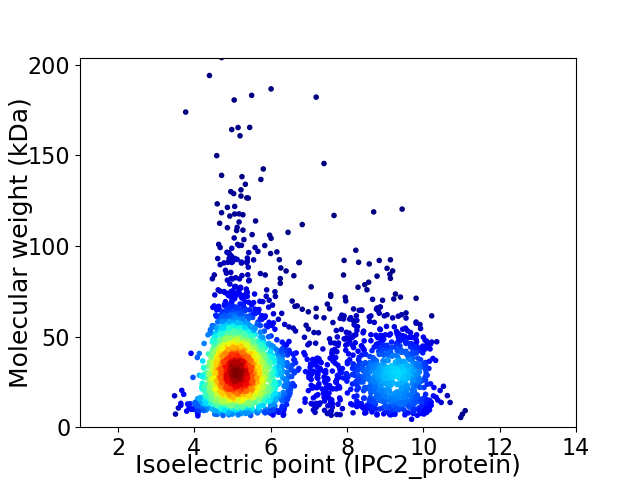
Virtual 2D-PAGE plot for 3046 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6PZR2|A0A0Q6PZR2_9MICO Uncharacterized protein OS=Leifsonia sp. Root112D2 OX=1736426 GN=ASC63_05435 PE=4 SV=1
MM1 pKa = 6.9VAILASLLVLASSLAPTLAAEE22 pKa = 4.42AAGPAVVAVTVSITDD37 pKa = 3.27PTGAVVSEE45 pKa = 4.23VDD47 pKa = 3.3ASSMANYY54 pKa = 9.4SVNVAYY60 pKa = 10.3SCSVADD66 pKa = 3.67CTGVQVVIDD75 pKa = 4.25PTVLDD80 pKa = 5.0PIYY83 pKa = 10.36GQYY86 pKa = 10.37RR87 pKa = 11.84KK88 pKa = 8.21EE89 pKa = 4.06TAVTFTPPFSPAPSFTGSLGAGYY112 pKa = 7.94TVDD115 pKa = 3.95LGTVAVGTGGSFKK128 pKa = 10.55LAYY131 pKa = 10.13RR132 pKa = 11.84IDD134 pKa = 3.42ARR136 pKa = 11.84YY137 pKa = 10.18SGPQPGSFFLDD148 pKa = 4.21GSPITARR155 pKa = 11.84ATVSASNADD164 pKa = 3.44SSAVGTASANWVSSIPAAPTARR186 pKa = 11.84LSAPASIRR194 pKa = 11.84TDD196 pKa = 3.31TPVTITAEE204 pKa = 4.17GDD206 pKa = 3.94SRR208 pKa = 11.84CLSYY212 pKa = 11.31NGAGVLLGIPWLTCAKK228 pKa = 10.41DD229 pKa = 3.36YY230 pKa = 7.63TAPVTLPADD239 pKa = 3.68AQYY242 pKa = 11.4VAGSGGSYY250 pKa = 10.36NAGTRR255 pKa = 11.84TVTFSQTGAAAAGGLAFNDD274 pKa = 3.16NTFQIVFPSDD284 pKa = 3.26AFPTSGDD291 pKa = 3.24GCVASEE297 pKa = 4.21VFSVTEE303 pKa = 3.73TMTYY307 pKa = 10.89LDD309 pKa = 3.8GTVRR313 pKa = 11.84TADD316 pKa = 3.72PASTAIKK323 pKa = 9.98VQNCTPFAKK332 pKa = 10.44GALSKK337 pKa = 10.96AIGGRR342 pKa = 11.84DD343 pKa = 3.1GGTPTDD349 pKa = 4.81AIVDD353 pKa = 3.7IPLTPGGVNNKK364 pKa = 7.76YY365 pKa = 9.1WQVEE369 pKa = 3.78ADD371 pKa = 3.46NQANVAGVATITDD384 pKa = 3.79ADD386 pKa = 4.38LGQDD390 pKa = 3.97DD391 pKa = 4.72MPITSIRR398 pKa = 11.84IVGGGPATVNYY409 pKa = 9.16TLDD412 pKa = 3.86DD413 pKa = 3.98ATTGSATVAQGATWTPPSGHH433 pKa = 6.67TIAGVSATSSLLVGPNSVEE452 pKa = 3.86SGTAVTRR459 pKa = 11.84FIIRR463 pKa = 11.84LNFSVHH469 pKa = 7.29DD470 pKa = 3.99GTTPGPRR477 pKa = 11.84TNTATASIAYY487 pKa = 8.26PDD489 pKa = 3.79QPEE492 pKa = 5.21LGTLDD497 pKa = 4.31LGSTTATATLQVNADD512 pKa = 3.42VTLNAGGPSVTATGTPIVGGTVGWKK537 pKa = 9.3MSGAMDD543 pKa = 3.98NVAAGANYY551 pKa = 9.21RR552 pKa = 11.84PQYY555 pKa = 11.05VFLAPKK561 pKa = 9.62DD562 pKa = 3.65WNITAGSAAVSGVPGMTYY580 pKa = 10.22DD581 pKa = 3.86YY582 pKa = 10.27KK583 pKa = 10.3TVTYY587 pKa = 10.63NGEE590 pKa = 3.93SYY592 pKa = 11.18DD593 pKa = 3.79AVLATWPAAVTDD605 pKa = 3.65VGKK608 pKa = 10.87FNLPTMTVDD617 pKa = 3.92TVPTGAATAGTNNQTAYY634 pKa = 11.2LFLGDD639 pKa = 3.99VNNGPVALYY648 pKa = 10.24YY649 pKa = 10.43PNAYY653 pKa = 8.74TDD655 pKa = 4.32ASDD658 pKa = 4.47FDD660 pKa = 4.88GDD662 pKa = 3.99GNATEE667 pKa = 4.26QYY669 pKa = 9.52SWRR672 pKa = 11.84SNTTSLAPSKK682 pKa = 10.7SIAVTKK688 pKa = 8.4QICHH692 pKa = 7.05PDD694 pKa = 3.16AEE696 pKa = 4.49QADD699 pKa = 4.27GCDD702 pKa = 3.67WISDD706 pKa = 3.82SNITVGVPPNATAIKK721 pKa = 10.45YY722 pKa = 9.62RR723 pKa = 11.84VTLANSGNVDD733 pKa = 4.0LNNVVAYY740 pKa = 10.23DD741 pKa = 3.43VLPYY745 pKa = 10.99KK746 pKa = 10.67NDD748 pKa = 3.35TGLTDD753 pKa = 4.11GTASTPRR760 pKa = 11.84GSTVQEE766 pKa = 3.95VLATVSDD773 pKa = 3.98VSAGVDD779 pKa = 3.46LSYY782 pKa = 11.3SGSTNPARR790 pKa = 11.84PQVYY794 pKa = 9.06TGTTDD799 pKa = 3.46ADD801 pKa = 3.5DD802 pKa = 3.54WGTTVTEE809 pKa = 4.43AKK811 pKa = 10.32AIRR814 pKa = 11.84AQVTSLAAGASVTFDD829 pKa = 3.57YY830 pKa = 10.36EE831 pKa = 3.92ASLVDD836 pKa = 3.51GTANLVACNSIAATAATLAPIEE858 pKa = 4.23PAAVCASTQAADD870 pKa = 4.46LEE872 pKa = 4.67ITAPAHH878 pKa = 7.13LPMQQGRR885 pKa = 11.84PGAADD890 pKa = 3.21FTVINHH896 pKa = 6.72GGSPLSPATVTAVIPAGFSVTDD918 pKa = 4.23LAPIGYY924 pKa = 9.25ACSDD928 pKa = 3.61AAGDD932 pKa = 3.97SAPIAGPVTLSCVTQKK948 pKa = 11.0PDD950 pKa = 2.95GTVRR954 pKa = 11.84NLSLDD959 pKa = 3.77APDD962 pKa = 3.94TLSIPVLVDD971 pKa = 3.29EE972 pKa = 5.4DD973 pKa = 3.95ATGPSCIDD981 pKa = 3.25ASVTGTLYY989 pKa = 11.13DD990 pKa = 4.6PDD992 pKa = 3.92LTNNDD997 pKa = 3.26TSFCAAVADD1006 pKa = 4.11STLPLITLDD1015 pKa = 3.5KK1016 pKa = 10.5TDD1018 pKa = 3.5NTTSVTVGEE1027 pKa = 4.18QTTYY1031 pKa = 10.64SLTVASHH1038 pKa = 6.71LSSEE1042 pKa = 4.28AVAGAVLTDD1051 pKa = 3.55TLPAGEE1057 pKa = 4.59TFVSASDD1064 pKa = 3.43GGTASGQTVTWNLAEE1079 pKa = 4.63LNPTGAPSADD1089 pKa = 3.38GSDD1092 pKa = 3.49TTGGSGAAITVTVTVQIDD1110 pKa = 3.8DD1111 pKa = 4.05GTQDD1115 pKa = 3.5TLFNTASVSAPDD1127 pKa = 3.85PAGGPTLTDD1136 pKa = 3.28GATDD1140 pKa = 3.42TDD1142 pKa = 3.78TVRR1145 pKa = 11.84NVFTDD1150 pKa = 4.22LDD1152 pKa = 4.0PAIATPQNQSINTPLSDD1169 pKa = 3.24IVTTTGASLDD1179 pKa = 4.02PASVAQNTAPTHH1191 pKa = 5.39GSITIDD1197 pKa = 3.31TTTGAVTYY1205 pKa = 9.75IPAPGYY1211 pKa = 10.51SGADD1215 pKa = 3.19SYY1217 pKa = 11.94EE1218 pKa = 3.91VTVCDD1223 pKa = 3.57TSSPAQCHH1231 pKa = 4.9VSTVTVAVGVNVVDD1245 pKa = 4.8AVNDD1249 pKa = 3.8TDD1251 pKa = 3.34ATTAVTPVTTDD1262 pKa = 2.86VTGNDD1267 pKa = 3.77TTQSGQPLAHH1277 pKa = 6.27PTVPTSPTHH1286 pKa = 6.04GSAVVNGDD1294 pKa = 3.1GTITYY1299 pKa = 8.39TPAAAWSGTDD1309 pKa = 2.83SYY1311 pKa = 11.64GYY1313 pKa = 8.25QLCDD1317 pKa = 2.9TSTPTPVCDD1326 pKa = 3.14TATVQVTVANVFLDD1340 pKa = 3.91ATPSIPTAQNAPVTTPLADD1359 pKa = 2.95IVTTTGLLLDD1369 pKa = 4.23PTQVTQEE1376 pKa = 3.97TAPAHH1381 pKa = 5.62GSIAIDD1387 pKa = 3.44PHH1389 pKa = 7.92SGAVTYY1395 pKa = 9.7TPTAGYY1401 pKa = 8.51TGDD1404 pKa = 4.06DD1405 pKa = 3.54SYY1407 pKa = 11.96AIQVCDD1413 pKa = 3.55TDD1415 pKa = 5.23GVCHH1419 pKa = 6.56NSPVSVSVGANVVVVQDD1436 pKa = 3.46ATIGTRR1442 pKa = 11.84VNTDD1446 pKa = 3.12ADD1448 pKa = 3.96PVDD1451 pKa = 4.06VLTTATSASGQPLGNPTITDD1471 pKa = 3.56QPGHH1475 pKa = 5.46GTVVVNADD1483 pKa = 3.27GTISYY1488 pKa = 10.36GPEE1491 pKa = 3.47TDD1493 pKa = 3.83YY1494 pKa = 11.64VGSDD1498 pKa = 2.71SFTYY1502 pKa = 8.08TVCDD1506 pKa = 3.38TGSPTQACDD1515 pKa = 2.7TGTITVTVVAIADD1528 pKa = 3.94LVSTKK1533 pKa = 10.12TLDD1536 pKa = 3.28TATVVPGLPVSYY1548 pKa = 9.0TAMLHH1553 pKa = 6.7NSGPWTATNVHH1564 pKa = 6.67SIDD1567 pKa = 4.96PIDD1570 pKa = 3.97KK1571 pKa = 10.16RR1572 pKa = 11.84VMHH1575 pKa = 7.01AKK1577 pKa = 8.04GTPDD1581 pKa = 2.99ASVPGGLCLTRR1592 pKa = 11.84PTTTADD1598 pKa = 3.71LARR1601 pKa = 11.84LNPASGPYY1609 pKa = 9.82TIDD1612 pKa = 3.81AYY1614 pKa = 10.08PQVVDD1619 pKa = 4.4CSYY1622 pKa = 11.18PSIPSGTTVHH1632 pKa = 6.47DD1633 pKa = 4.79TITGTVDD1640 pKa = 3.29PAALPGGTILNQAPVYY1656 pKa = 10.45ADD1658 pKa = 4.12SYY1660 pKa = 12.08DD1661 pKa = 3.8PDD1663 pKa = 3.51LSNNVGTATATITAPADD1680 pKa = 3.42VDD1682 pKa = 3.91PTTSAPSGLARR1693 pKa = 11.84TGVDD1697 pKa = 3.26AAPTGILALLLMLAGAGTLFLARR1720 pKa = 11.84RR1721 pKa = 11.84RR1722 pKa = 11.84RR1723 pKa = 11.84RR1724 pKa = 11.84AA1725 pKa = 2.97
MM1 pKa = 6.9VAILASLLVLASSLAPTLAAEE22 pKa = 4.42AAGPAVVAVTVSITDD37 pKa = 3.27PTGAVVSEE45 pKa = 4.23VDD47 pKa = 3.3ASSMANYY54 pKa = 9.4SVNVAYY60 pKa = 10.3SCSVADD66 pKa = 3.67CTGVQVVIDD75 pKa = 4.25PTVLDD80 pKa = 5.0PIYY83 pKa = 10.36GQYY86 pKa = 10.37RR87 pKa = 11.84KK88 pKa = 8.21EE89 pKa = 4.06TAVTFTPPFSPAPSFTGSLGAGYY112 pKa = 7.94TVDD115 pKa = 3.95LGTVAVGTGGSFKK128 pKa = 10.55LAYY131 pKa = 10.13RR132 pKa = 11.84IDD134 pKa = 3.42ARR136 pKa = 11.84YY137 pKa = 10.18SGPQPGSFFLDD148 pKa = 4.21GSPITARR155 pKa = 11.84ATVSASNADD164 pKa = 3.44SSAVGTASANWVSSIPAAPTARR186 pKa = 11.84LSAPASIRR194 pKa = 11.84TDD196 pKa = 3.31TPVTITAEE204 pKa = 4.17GDD206 pKa = 3.94SRR208 pKa = 11.84CLSYY212 pKa = 11.31NGAGVLLGIPWLTCAKK228 pKa = 10.41DD229 pKa = 3.36YY230 pKa = 7.63TAPVTLPADD239 pKa = 3.68AQYY242 pKa = 11.4VAGSGGSYY250 pKa = 10.36NAGTRR255 pKa = 11.84TVTFSQTGAAAAGGLAFNDD274 pKa = 3.16NTFQIVFPSDD284 pKa = 3.26AFPTSGDD291 pKa = 3.24GCVASEE297 pKa = 4.21VFSVTEE303 pKa = 3.73TMTYY307 pKa = 10.89LDD309 pKa = 3.8GTVRR313 pKa = 11.84TADD316 pKa = 3.72PASTAIKK323 pKa = 9.98VQNCTPFAKK332 pKa = 10.44GALSKK337 pKa = 10.96AIGGRR342 pKa = 11.84DD343 pKa = 3.1GGTPTDD349 pKa = 4.81AIVDD353 pKa = 3.7IPLTPGGVNNKK364 pKa = 7.76YY365 pKa = 9.1WQVEE369 pKa = 3.78ADD371 pKa = 3.46NQANVAGVATITDD384 pKa = 3.79ADD386 pKa = 4.38LGQDD390 pKa = 3.97DD391 pKa = 4.72MPITSIRR398 pKa = 11.84IVGGGPATVNYY409 pKa = 9.16TLDD412 pKa = 3.86DD413 pKa = 3.98ATTGSATVAQGATWTPPSGHH433 pKa = 6.67TIAGVSATSSLLVGPNSVEE452 pKa = 3.86SGTAVTRR459 pKa = 11.84FIIRR463 pKa = 11.84LNFSVHH469 pKa = 7.29DD470 pKa = 3.99GTTPGPRR477 pKa = 11.84TNTATASIAYY487 pKa = 8.26PDD489 pKa = 3.79QPEE492 pKa = 5.21LGTLDD497 pKa = 4.31LGSTTATATLQVNADD512 pKa = 3.42VTLNAGGPSVTATGTPIVGGTVGWKK537 pKa = 9.3MSGAMDD543 pKa = 3.98NVAAGANYY551 pKa = 9.21RR552 pKa = 11.84PQYY555 pKa = 11.05VFLAPKK561 pKa = 9.62DD562 pKa = 3.65WNITAGSAAVSGVPGMTYY580 pKa = 10.22DD581 pKa = 3.86YY582 pKa = 10.27KK583 pKa = 10.3TVTYY587 pKa = 10.63NGEE590 pKa = 3.93SYY592 pKa = 11.18DD593 pKa = 3.79AVLATWPAAVTDD605 pKa = 3.65VGKK608 pKa = 10.87FNLPTMTVDD617 pKa = 3.92TVPTGAATAGTNNQTAYY634 pKa = 11.2LFLGDD639 pKa = 3.99VNNGPVALYY648 pKa = 10.24YY649 pKa = 10.43PNAYY653 pKa = 8.74TDD655 pKa = 4.32ASDD658 pKa = 4.47FDD660 pKa = 4.88GDD662 pKa = 3.99GNATEE667 pKa = 4.26QYY669 pKa = 9.52SWRR672 pKa = 11.84SNTTSLAPSKK682 pKa = 10.7SIAVTKK688 pKa = 8.4QICHH692 pKa = 7.05PDD694 pKa = 3.16AEE696 pKa = 4.49QADD699 pKa = 4.27GCDD702 pKa = 3.67WISDD706 pKa = 3.82SNITVGVPPNATAIKK721 pKa = 10.45YY722 pKa = 9.62RR723 pKa = 11.84VTLANSGNVDD733 pKa = 4.0LNNVVAYY740 pKa = 10.23DD741 pKa = 3.43VLPYY745 pKa = 10.99KK746 pKa = 10.67NDD748 pKa = 3.35TGLTDD753 pKa = 4.11GTASTPRR760 pKa = 11.84GSTVQEE766 pKa = 3.95VLATVSDD773 pKa = 3.98VSAGVDD779 pKa = 3.46LSYY782 pKa = 11.3SGSTNPARR790 pKa = 11.84PQVYY794 pKa = 9.06TGTTDD799 pKa = 3.46ADD801 pKa = 3.5DD802 pKa = 3.54WGTTVTEE809 pKa = 4.43AKK811 pKa = 10.32AIRR814 pKa = 11.84AQVTSLAAGASVTFDD829 pKa = 3.57YY830 pKa = 10.36EE831 pKa = 3.92ASLVDD836 pKa = 3.51GTANLVACNSIAATAATLAPIEE858 pKa = 4.23PAAVCASTQAADD870 pKa = 4.46LEE872 pKa = 4.67ITAPAHH878 pKa = 7.13LPMQQGRR885 pKa = 11.84PGAADD890 pKa = 3.21FTVINHH896 pKa = 6.72GGSPLSPATVTAVIPAGFSVTDD918 pKa = 4.23LAPIGYY924 pKa = 9.25ACSDD928 pKa = 3.61AAGDD932 pKa = 3.97SAPIAGPVTLSCVTQKK948 pKa = 11.0PDD950 pKa = 2.95GTVRR954 pKa = 11.84NLSLDD959 pKa = 3.77APDD962 pKa = 3.94TLSIPVLVDD971 pKa = 3.29EE972 pKa = 5.4DD973 pKa = 3.95ATGPSCIDD981 pKa = 3.25ASVTGTLYY989 pKa = 11.13DD990 pKa = 4.6PDD992 pKa = 3.92LTNNDD997 pKa = 3.26TSFCAAVADD1006 pKa = 4.11STLPLITLDD1015 pKa = 3.5KK1016 pKa = 10.5TDD1018 pKa = 3.5NTTSVTVGEE1027 pKa = 4.18QTTYY1031 pKa = 10.64SLTVASHH1038 pKa = 6.71LSSEE1042 pKa = 4.28AVAGAVLTDD1051 pKa = 3.55TLPAGEE1057 pKa = 4.59TFVSASDD1064 pKa = 3.43GGTASGQTVTWNLAEE1079 pKa = 4.63LNPTGAPSADD1089 pKa = 3.38GSDD1092 pKa = 3.49TTGGSGAAITVTVTVQIDD1110 pKa = 3.8DD1111 pKa = 4.05GTQDD1115 pKa = 3.5TLFNTASVSAPDD1127 pKa = 3.85PAGGPTLTDD1136 pKa = 3.28GATDD1140 pKa = 3.42TDD1142 pKa = 3.78TVRR1145 pKa = 11.84NVFTDD1150 pKa = 4.22LDD1152 pKa = 4.0PAIATPQNQSINTPLSDD1169 pKa = 3.24IVTTTGASLDD1179 pKa = 4.02PASVAQNTAPTHH1191 pKa = 5.39GSITIDD1197 pKa = 3.31TTTGAVTYY1205 pKa = 9.75IPAPGYY1211 pKa = 10.51SGADD1215 pKa = 3.19SYY1217 pKa = 11.94EE1218 pKa = 3.91VTVCDD1223 pKa = 3.57TSSPAQCHH1231 pKa = 4.9VSTVTVAVGVNVVDD1245 pKa = 4.8AVNDD1249 pKa = 3.8TDD1251 pKa = 3.34ATTAVTPVTTDD1262 pKa = 2.86VTGNDD1267 pKa = 3.77TTQSGQPLAHH1277 pKa = 6.27PTVPTSPTHH1286 pKa = 6.04GSAVVNGDD1294 pKa = 3.1GTITYY1299 pKa = 8.39TPAAAWSGTDD1309 pKa = 2.83SYY1311 pKa = 11.64GYY1313 pKa = 8.25QLCDD1317 pKa = 2.9TSTPTPVCDD1326 pKa = 3.14TATVQVTVANVFLDD1340 pKa = 3.91ATPSIPTAQNAPVTTPLADD1359 pKa = 2.95IVTTTGLLLDD1369 pKa = 4.23PTQVTQEE1376 pKa = 3.97TAPAHH1381 pKa = 5.62GSIAIDD1387 pKa = 3.44PHH1389 pKa = 7.92SGAVTYY1395 pKa = 9.7TPTAGYY1401 pKa = 8.51TGDD1404 pKa = 4.06DD1405 pKa = 3.54SYY1407 pKa = 11.96AIQVCDD1413 pKa = 3.55TDD1415 pKa = 5.23GVCHH1419 pKa = 6.56NSPVSVSVGANVVVVQDD1436 pKa = 3.46ATIGTRR1442 pKa = 11.84VNTDD1446 pKa = 3.12ADD1448 pKa = 3.96PVDD1451 pKa = 4.06VLTTATSASGQPLGNPTITDD1471 pKa = 3.56QPGHH1475 pKa = 5.46GTVVVNADD1483 pKa = 3.27GTISYY1488 pKa = 10.36GPEE1491 pKa = 3.47TDD1493 pKa = 3.83YY1494 pKa = 11.64VGSDD1498 pKa = 2.71SFTYY1502 pKa = 8.08TVCDD1506 pKa = 3.38TGSPTQACDD1515 pKa = 2.7TGTITVTVVAIADD1528 pKa = 3.94LVSTKK1533 pKa = 10.12TLDD1536 pKa = 3.28TATVVPGLPVSYY1548 pKa = 9.0TAMLHH1553 pKa = 6.7NSGPWTATNVHH1564 pKa = 6.67SIDD1567 pKa = 4.96PIDD1570 pKa = 3.97KK1571 pKa = 10.16RR1572 pKa = 11.84VMHH1575 pKa = 7.01AKK1577 pKa = 8.04GTPDD1581 pKa = 2.99ASVPGGLCLTRR1592 pKa = 11.84PTTTADD1598 pKa = 3.71LARR1601 pKa = 11.84LNPASGPYY1609 pKa = 9.82TIDD1612 pKa = 3.81AYY1614 pKa = 10.08PQVVDD1619 pKa = 4.4CSYY1622 pKa = 11.18PSIPSGTTVHH1632 pKa = 6.47DD1633 pKa = 4.79TITGTVDD1640 pKa = 3.29PAALPGGTILNQAPVYY1656 pKa = 10.45ADD1658 pKa = 4.12SYY1660 pKa = 12.08DD1661 pKa = 3.8PDD1663 pKa = 3.51LSNNVGTATATITAPADD1680 pKa = 3.42VDD1682 pKa = 3.91PTTSAPSGLARR1693 pKa = 11.84TGVDD1697 pKa = 3.26AAPTGILALLLMLAGAGTLFLARR1720 pKa = 11.84RR1721 pKa = 11.84RR1722 pKa = 11.84RR1723 pKa = 11.84RR1724 pKa = 11.84AA1725 pKa = 2.97
Molecular weight: 173.94 kDa
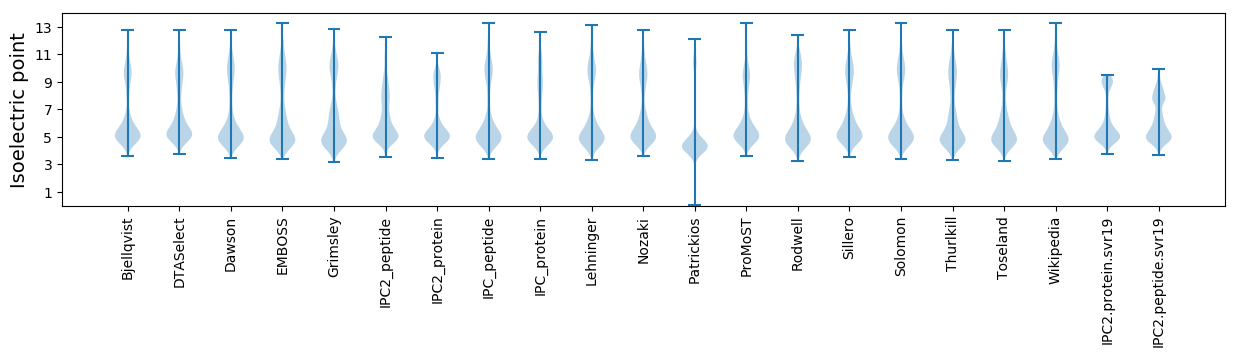
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6Q0S2|A0A0Q6Q0S2_9MICO Beta-glucosidase OS=Leifsonia sp. Root112D2 OX=1736426 GN=ASC63_01865 PE=3 SV=1
MM1 pKa = 7.33KK2 pKa = 10.29RR3 pKa = 11.84IWPLLVPGVILSAVGLVWTLQGLNVLRR30 pKa = 11.84GSVMSGSSLWATMGPIVLLLGLVLIAIAIARR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.58RR66 pKa = 3.2
MM1 pKa = 7.33KK2 pKa = 10.29RR3 pKa = 11.84IWPLLVPGVILSAVGLVWTLQGLNVLRR30 pKa = 11.84GSVMSGSSLWATMGPIVLLLGLVLIAIAIARR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.58RR66 pKa = 3.2
Molecular weight: 7.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1001125 |
37 |
1981 |
328.7 |
35.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.05 ± 0.063 | 0.484 ± 0.01 |
5.723 ± 0.035 | 5.381 ± 0.048 |
3.232 ± 0.031 | 8.747 ± 0.036 |
2.024 ± 0.021 | 4.913 ± 0.032 |
2.445 ± 0.035 | 10.223 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.874 ± 0.018 | 2.411 ± 0.023 |
5.089 ± 0.033 | 2.981 ± 0.022 |
6.784 ± 0.046 | 6.172 ± 0.032 |
6.192 ± 0.043 | 8.729 ± 0.036 |
1.445 ± 0.016 | 2.102 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
