
Serinibacter salmoneus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Beutenbergiaceae; Serinibacter
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
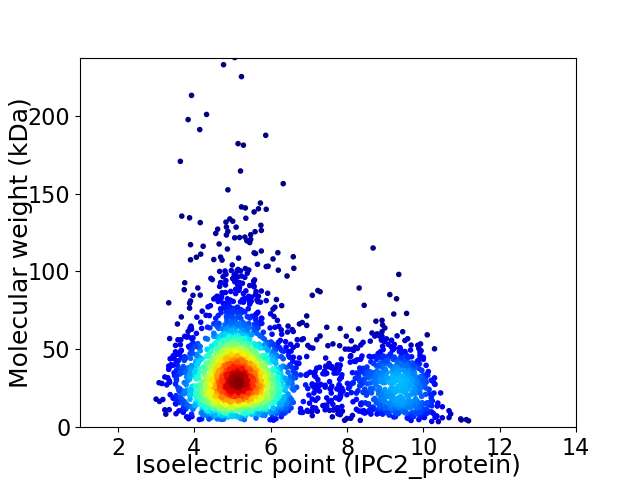
Virtual 2D-PAGE plot for 2863 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9D1Y5|A0A2A9D1Y5_9MICO Transcriptional regulator AbiEi antitoxin Type IV TA system OS=Serinibacter salmoneus OX=556530 GN=ATL40_2294 PE=4 SV=1
MM1 pKa = 7.55SSPRR5 pKa = 11.84ATTRR9 pKa = 11.84IRR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84PALIAACTALLALATACTPEE34 pKa = 6.0DD35 pKa = 4.47GEE37 pKa = 5.02SPDD40 pKa = 4.19PTPSAAPEE48 pKa = 4.12DD49 pKa = 4.0ATTDD53 pKa = 3.51GSITGDD59 pKa = 3.47LTLSDD64 pKa = 5.21LGPDD68 pKa = 3.25GDD70 pKa = 4.04TTVFPGGSAQAAVDD84 pKa = 3.59QVEE87 pKa = 4.25QVVDD91 pKa = 3.62QVLADD96 pKa = 3.6TGVPGAAVAIVTGDD110 pKa = 3.27EE111 pKa = 3.92VLYY114 pKa = 10.42TGGFGVRR121 pKa = 11.84DD122 pKa = 3.94LTTGEE127 pKa = 4.19PVDD130 pKa = 4.14AEE132 pKa = 4.65TVFQIASVSKK142 pKa = 10.3SVGATVVATQVTDD155 pKa = 4.91GVVSWDD161 pKa = 4.38DD162 pKa = 3.46PSHH165 pKa = 6.38TYY167 pKa = 10.61LPEE170 pKa = 6.14LEE172 pKa = 5.78LSDD175 pKa = 3.77AWVSEE180 pKa = 3.91HH181 pKa = 5.53VTIGDD186 pKa = 4.4LYY188 pKa = 9.69SHH190 pKa = 6.68RR191 pKa = 11.84TGLPHH196 pKa = 7.55AAGDD200 pKa = 3.87LLEE203 pKa = 5.72DD204 pKa = 3.03IGYY207 pKa = 10.04DD208 pKa = 2.94RR209 pKa = 11.84TEE211 pKa = 3.44ILQRR215 pKa = 11.84LVYY218 pKa = 10.41QPLNPFRR225 pKa = 11.84TSYY228 pKa = 11.28AYY230 pKa = 10.53ANYY233 pKa = 7.83GTTAGGEE240 pKa = 4.36AVAHH244 pKa = 6.11AAGTTWEE251 pKa = 5.04DD252 pKa = 3.31LSQDD256 pKa = 3.41ALYY259 pKa = 10.73DD260 pKa = 3.93PLGMTSTSSRR270 pKa = 11.84HH271 pKa = 4.5VDD273 pKa = 3.18YY274 pKa = 10.95EE275 pKa = 3.94AAANRR280 pKa = 11.84ATLHH284 pKa = 6.18MYY286 pKa = 10.68LGDD289 pKa = 3.84GEE291 pKa = 4.61FEE293 pKa = 4.36HH294 pKa = 7.27VAEE297 pKa = 5.25RR298 pKa = 11.84DD299 pKa = 3.61PDD301 pKa = 3.87PQSPAGGVSSTASDD315 pKa = 3.67LALWLQLLLNDD326 pKa = 4.07GVGPQGQEE334 pKa = 3.93LIAPEE339 pKa = 4.33ALQPAITPQSFSAPASVPQARR360 pKa = 11.84SGSYY364 pKa = 10.45GYY366 pKa = 10.66GFNVGVTAGGQPKK379 pKa = 9.93LSHH382 pKa = 6.46SGAFVIGTGTSFVVLPGLDD401 pKa = 3.45VAVVALTNGGPVGAAEE417 pKa = 5.62AITASLADD425 pKa = 3.65LAEE428 pKa = 4.44YY429 pKa = 11.3GEE431 pKa = 4.42VTRR434 pKa = 11.84DD435 pKa = 2.76WAAGYY440 pKa = 9.06GALFEE445 pKa = 5.33PYY447 pKa = 9.07HH448 pKa = 7.1APVGDD453 pKa = 4.53LAEE456 pKa = 4.22QDD458 pKa = 3.98PPADD462 pKa = 3.21AAAPQDD468 pKa = 3.44LATYY472 pKa = 8.6TGAWEE477 pKa = 3.63NDD479 pKa = 3.59YY480 pKa = 11.25YY481 pKa = 11.61GEE483 pKa = 4.24AVVSVAGEE491 pKa = 4.04TLVAEE496 pKa = 5.01LGPDD500 pKa = 3.1GGYY503 pKa = 10.41VLEE506 pKa = 4.26LQPWDD511 pKa = 3.34GDD513 pKa = 3.65TFAFVPTGEE522 pKa = 4.05NAPLGSLSSATFTVPDD538 pKa = 3.63GTARR542 pKa = 11.84AEE544 pKa = 4.14TMTLEE549 pKa = 4.35FFDD552 pKa = 3.94ATGLGTWTRR561 pKa = 11.84ANN563 pKa = 3.4
MM1 pKa = 7.55SSPRR5 pKa = 11.84ATTRR9 pKa = 11.84IRR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84PALIAACTALLALATACTPEE34 pKa = 6.0DD35 pKa = 4.47GEE37 pKa = 5.02SPDD40 pKa = 4.19PTPSAAPEE48 pKa = 4.12DD49 pKa = 4.0ATTDD53 pKa = 3.51GSITGDD59 pKa = 3.47LTLSDD64 pKa = 5.21LGPDD68 pKa = 3.25GDD70 pKa = 4.04TTVFPGGSAQAAVDD84 pKa = 3.59QVEE87 pKa = 4.25QVVDD91 pKa = 3.62QVLADD96 pKa = 3.6TGVPGAAVAIVTGDD110 pKa = 3.27EE111 pKa = 3.92VLYY114 pKa = 10.42TGGFGVRR121 pKa = 11.84DD122 pKa = 3.94LTTGEE127 pKa = 4.19PVDD130 pKa = 4.14AEE132 pKa = 4.65TVFQIASVSKK142 pKa = 10.3SVGATVVATQVTDD155 pKa = 4.91GVVSWDD161 pKa = 4.38DD162 pKa = 3.46PSHH165 pKa = 6.38TYY167 pKa = 10.61LPEE170 pKa = 6.14LEE172 pKa = 5.78LSDD175 pKa = 3.77AWVSEE180 pKa = 3.91HH181 pKa = 5.53VTIGDD186 pKa = 4.4LYY188 pKa = 9.69SHH190 pKa = 6.68RR191 pKa = 11.84TGLPHH196 pKa = 7.55AAGDD200 pKa = 3.87LLEE203 pKa = 5.72DD204 pKa = 3.03IGYY207 pKa = 10.04DD208 pKa = 2.94RR209 pKa = 11.84TEE211 pKa = 3.44ILQRR215 pKa = 11.84LVYY218 pKa = 10.41QPLNPFRR225 pKa = 11.84TSYY228 pKa = 11.28AYY230 pKa = 10.53ANYY233 pKa = 7.83GTTAGGEE240 pKa = 4.36AVAHH244 pKa = 6.11AAGTTWEE251 pKa = 5.04DD252 pKa = 3.31LSQDD256 pKa = 3.41ALYY259 pKa = 10.73DD260 pKa = 3.93PLGMTSTSSRR270 pKa = 11.84HH271 pKa = 4.5VDD273 pKa = 3.18YY274 pKa = 10.95EE275 pKa = 3.94AAANRR280 pKa = 11.84ATLHH284 pKa = 6.18MYY286 pKa = 10.68LGDD289 pKa = 3.84GEE291 pKa = 4.61FEE293 pKa = 4.36HH294 pKa = 7.27VAEE297 pKa = 5.25RR298 pKa = 11.84DD299 pKa = 3.61PDD301 pKa = 3.87PQSPAGGVSSTASDD315 pKa = 3.67LALWLQLLLNDD326 pKa = 4.07GVGPQGQEE334 pKa = 3.93LIAPEE339 pKa = 4.33ALQPAITPQSFSAPASVPQARR360 pKa = 11.84SGSYY364 pKa = 10.45GYY366 pKa = 10.66GFNVGVTAGGQPKK379 pKa = 9.93LSHH382 pKa = 6.46SGAFVIGTGTSFVVLPGLDD401 pKa = 3.45VAVVALTNGGPVGAAEE417 pKa = 5.62AITASLADD425 pKa = 3.65LAEE428 pKa = 4.44YY429 pKa = 11.3GEE431 pKa = 4.42VTRR434 pKa = 11.84DD435 pKa = 2.76WAAGYY440 pKa = 9.06GALFEE445 pKa = 5.33PYY447 pKa = 9.07HH448 pKa = 7.1APVGDD453 pKa = 4.53LAEE456 pKa = 4.22QDD458 pKa = 3.98PPADD462 pKa = 3.21AAAPQDD468 pKa = 3.44LATYY472 pKa = 8.6TGAWEE477 pKa = 3.63NDD479 pKa = 3.59YY480 pKa = 11.25YY481 pKa = 11.61GEE483 pKa = 4.24AVVSVAGEE491 pKa = 4.04TLVAEE496 pKa = 5.01LGPDD500 pKa = 3.1GGYY503 pKa = 10.41VLEE506 pKa = 4.26LQPWDD511 pKa = 3.34GDD513 pKa = 3.65TFAFVPTGEE522 pKa = 4.05NAPLGSLSSATFTVPDD538 pKa = 3.63GTARR542 pKa = 11.84AEE544 pKa = 4.14TMTLEE549 pKa = 4.35FFDD552 pKa = 3.94ATGLGTWTRR561 pKa = 11.84ANN563 pKa = 3.4
Molecular weight: 58.18 kDa
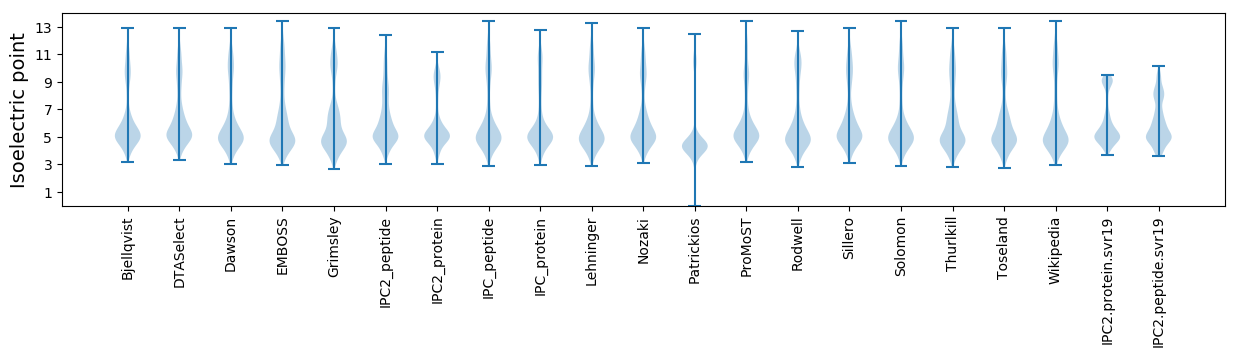
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9D417|A0A2A9D417_9MICO DNA invertase Pin-like site-specific DNA recombinase OS=Serinibacter salmoneus OX=556530 GN=ATL40_2312 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.64HH17 pKa = 5.77RR18 pKa = 11.84KK19 pKa = 7.85LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84HH26 pKa = 3.6QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.64HH17 pKa = 5.77RR18 pKa = 11.84KK19 pKa = 7.85LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84HH26 pKa = 3.6QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
984522 |
32 |
2222 |
343.9 |
36.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.292 ± 0.069 | 0.552 ± 0.01 |
5.786 ± 0.04 | 6.147 ± 0.053 |
2.519 ± 0.028 | 9.25 ± 0.04 |
2.097 ± 0.025 | 3.727 ± 0.028 |
1.385 ± 0.026 | 10.27 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.836 ± 0.019 | 1.595 ± 0.022 |
5.772 ± 0.033 | 2.895 ± 0.027 |
7.595 ± 0.051 | 5.454 ± 0.033 |
6.341 ± 0.043 | 9.082 ± 0.047 |
1.549 ± 0.023 | 1.857 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
