
Leucobacter sp. UCD-THU
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Leucobacter; unclassified Leucobacter
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
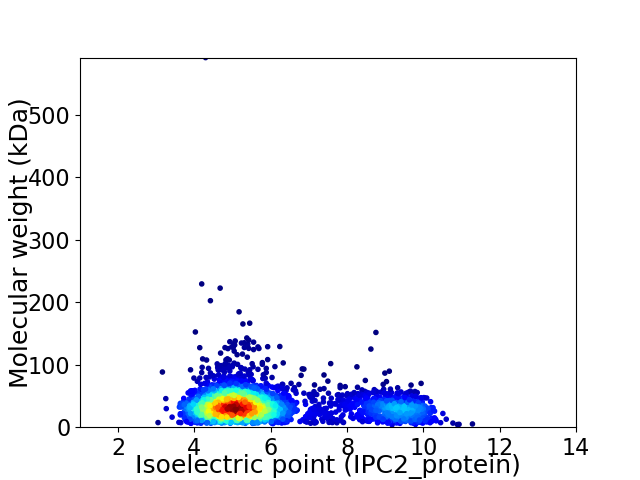
Virtual 2D-PAGE plot for 2878 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A061LXI6|A0A061LXI6_9MICO Glutamine synthetase OS=Leucobacter sp. UCD-THU OX=1292023 GN=H490_0112870 PE=3 SV=1
MM1 pKa = 7.82SSRR4 pKa = 11.84TVSARR9 pKa = 11.84AALPAILAASLLTLVACSGSGGGKK33 pKa = 9.25NASDD37 pKa = 3.99DD38 pKa = 3.91EE39 pKa = 5.22GPLSEE44 pKa = 4.64YY45 pKa = 10.85LSSLSSAEE53 pKa = 4.14TEE55 pKa = 3.81SEE57 pKa = 3.97EE58 pKa = 4.14FYY60 pKa = 11.4EE61 pKa = 4.57RR62 pKa = 11.84INAKK66 pKa = 10.14RR67 pKa = 11.84EE68 pKa = 3.92EE69 pKa = 4.2LVAEE73 pKa = 4.49CMAGEE78 pKa = 4.39GFDD81 pKa = 3.92YY82 pKa = 10.98QPNSQNGGTSTLGDD96 pKa = 4.63DD97 pKa = 4.45GIEE100 pKa = 3.92WGSLEE105 pKa = 4.08FAEE108 pKa = 4.38QYY110 pKa = 11.16GYY112 pKa = 11.36GIVDD116 pKa = 3.47WPGLAEE122 pKa = 4.22MEE124 pKa = 4.41QQSEE128 pKa = 5.08EE129 pKa = 4.24YY130 pKa = 10.6VDD132 pKa = 4.05PNQGYY137 pKa = 9.1VEE139 pKa = 4.29SLSEE143 pKa = 4.23SEE145 pKa = 3.86QQAYY149 pKa = 10.12YY150 pKa = 10.1EE151 pKa = 4.67ALSGPQPSEE160 pKa = 3.48EE161 pKa = 4.4DD162 pKa = 3.14MAAMEE167 pKa = 4.83DD168 pKa = 5.02GAYY171 pKa = 9.7EE172 pKa = 4.05WDD174 pKa = 3.43WTQSGCYY181 pKa = 9.13GAADD185 pKa = 3.89HH186 pKa = 6.73EE187 pKa = 4.72VQDD190 pKa = 4.41EE191 pKa = 4.52YY192 pKa = 11.55GSAMAAYY199 pKa = 7.87EE200 pKa = 4.12DD201 pKa = 4.41PEE203 pKa = 3.99FAEE206 pKa = 5.16LFEE209 pKa = 4.77SMNAVWSEE217 pKa = 3.83LYY219 pKa = 10.9NYY221 pKa = 9.0DD222 pKa = 4.87DD223 pKa = 5.28PGAPSPNDD231 pKa = 3.37DD232 pKa = 3.35VAKK235 pKa = 10.59LDD237 pKa = 4.2RR238 pKa = 11.84EE239 pKa = 4.21WSEE242 pKa = 4.2CMADD246 pKa = 4.14AGYY249 pKa = 9.08TDD251 pKa = 4.88LVSPQTAMDD260 pKa = 4.13GLNEE264 pKa = 4.07EE265 pKa = 4.73WNTLQTPPEE274 pKa = 4.21GTGDD278 pKa = 4.82DD279 pKa = 3.65LAGWSGPSDD288 pKa = 3.44RR289 pKa = 11.84QKK291 pKa = 11.3SEE293 pKa = 3.65FQQRR297 pKa = 11.84EE298 pKa = 3.5IEE300 pKa = 4.24MAVTDD305 pKa = 4.55WKK307 pKa = 11.17CKK309 pKa = 10.52DD310 pKa = 3.58GIDD313 pKa = 3.9YY314 pKa = 7.68EE315 pKa = 4.48QQVRR319 pKa = 11.84TVQFDD324 pKa = 3.66LEE326 pKa = 4.11QRR328 pKa = 11.84FVDD331 pKa = 3.5EE332 pKa = 5.35HH333 pKa = 6.25RR334 pKa = 11.84AEE336 pKa = 4.35LDD338 pKa = 3.22ALIAKK343 pKa = 9.96HH344 pKa = 6.41GADD347 pKa = 3.29QKK349 pKa = 11.46KK350 pKa = 10.08SS351 pKa = 3.35
MM1 pKa = 7.82SSRR4 pKa = 11.84TVSARR9 pKa = 11.84AALPAILAASLLTLVACSGSGGGKK33 pKa = 9.25NASDD37 pKa = 3.99DD38 pKa = 3.91EE39 pKa = 5.22GPLSEE44 pKa = 4.64YY45 pKa = 10.85LSSLSSAEE53 pKa = 4.14TEE55 pKa = 3.81SEE57 pKa = 3.97EE58 pKa = 4.14FYY60 pKa = 11.4EE61 pKa = 4.57RR62 pKa = 11.84INAKK66 pKa = 10.14RR67 pKa = 11.84EE68 pKa = 3.92EE69 pKa = 4.2LVAEE73 pKa = 4.49CMAGEE78 pKa = 4.39GFDD81 pKa = 3.92YY82 pKa = 10.98QPNSQNGGTSTLGDD96 pKa = 4.63DD97 pKa = 4.45GIEE100 pKa = 3.92WGSLEE105 pKa = 4.08FAEE108 pKa = 4.38QYY110 pKa = 11.16GYY112 pKa = 11.36GIVDD116 pKa = 3.47WPGLAEE122 pKa = 4.22MEE124 pKa = 4.41QQSEE128 pKa = 5.08EE129 pKa = 4.24YY130 pKa = 10.6VDD132 pKa = 4.05PNQGYY137 pKa = 9.1VEE139 pKa = 4.29SLSEE143 pKa = 4.23SEE145 pKa = 3.86QQAYY149 pKa = 10.12YY150 pKa = 10.1EE151 pKa = 4.67ALSGPQPSEE160 pKa = 3.48EE161 pKa = 4.4DD162 pKa = 3.14MAAMEE167 pKa = 4.83DD168 pKa = 5.02GAYY171 pKa = 9.7EE172 pKa = 4.05WDD174 pKa = 3.43WTQSGCYY181 pKa = 9.13GAADD185 pKa = 3.89HH186 pKa = 6.73EE187 pKa = 4.72VQDD190 pKa = 4.41EE191 pKa = 4.52YY192 pKa = 11.55GSAMAAYY199 pKa = 7.87EE200 pKa = 4.12DD201 pKa = 4.41PEE203 pKa = 3.99FAEE206 pKa = 5.16LFEE209 pKa = 4.77SMNAVWSEE217 pKa = 3.83LYY219 pKa = 10.9NYY221 pKa = 9.0DD222 pKa = 4.87DD223 pKa = 5.28PGAPSPNDD231 pKa = 3.37DD232 pKa = 3.35VAKK235 pKa = 10.59LDD237 pKa = 4.2RR238 pKa = 11.84EE239 pKa = 4.21WSEE242 pKa = 4.2CMADD246 pKa = 4.14AGYY249 pKa = 9.08TDD251 pKa = 4.88LVSPQTAMDD260 pKa = 4.13GLNEE264 pKa = 4.07EE265 pKa = 4.73WNTLQTPPEE274 pKa = 4.21GTGDD278 pKa = 4.82DD279 pKa = 3.65LAGWSGPSDD288 pKa = 3.44RR289 pKa = 11.84QKK291 pKa = 11.3SEE293 pKa = 3.65FQQRR297 pKa = 11.84EE298 pKa = 3.5IEE300 pKa = 4.24MAVTDD305 pKa = 4.55WKK307 pKa = 11.17CKK309 pKa = 10.52DD310 pKa = 3.58GIDD313 pKa = 3.9YY314 pKa = 7.68EE315 pKa = 4.48QQVRR319 pKa = 11.84TVQFDD324 pKa = 3.66LEE326 pKa = 4.11QRR328 pKa = 11.84FVDD331 pKa = 3.5EE332 pKa = 5.35HH333 pKa = 6.25RR334 pKa = 11.84AEE336 pKa = 4.35LDD338 pKa = 3.22ALIAKK343 pKa = 9.96HH344 pKa = 6.41GADD347 pKa = 3.29QKK349 pKa = 11.46KK350 pKa = 10.08SS351 pKa = 3.35
Molecular weight: 38.69 kDa
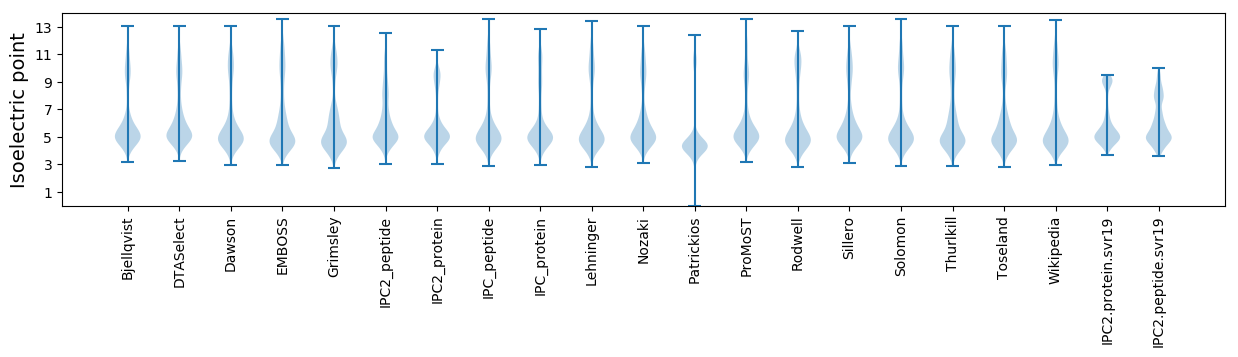
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A061LYY7|A0A061LYY7_9MICO Phosphatidylglycerol--prolipoprotein diacylglyceryl transferase OS=Leucobacter sp. UCD-THU OX=1292023 GN=lgt PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.45GRR40 pKa = 11.84SKK42 pKa = 10.13LTAA45 pKa = 4.04
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.45GRR40 pKa = 11.84SKK42 pKa = 10.13LTAA45 pKa = 4.04
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
968228 |
36 |
5399 |
336.4 |
35.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.622 ± 0.082 | 0.545 ± 0.01 |
5.8 ± 0.045 | 6.392 ± 0.045 |
3.126 ± 0.03 | 9.293 ± 0.041 |
1.947 ± 0.02 | 4.538 ± 0.033 |
1.87 ± 0.034 | 10.292 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.815 ± 0.017 | 1.872 ± 0.026 |
5.476 ± 0.031 | 2.994 ± 0.026 |
7.579 ± 0.053 | 5.578 ± 0.028 |
5.474 ± 0.038 | 8.496 ± 0.039 |
1.409 ± 0.021 | 1.88 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
