
Achromatium sp. WMS3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Thiotrichaceae; Achromatium; unclassified Achromatium
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
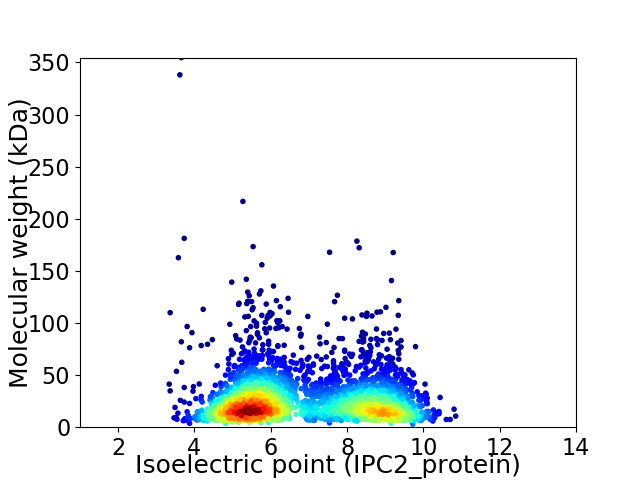
Virtual 2D-PAGE plot for 3099 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M1JBI1|A0A0M1JBI1_9GAMM Glutamine-synthetase adenylyltransferase (Fragment) OS=Achromatium sp. WMS3 OX=1604836 GN=TI05_13615 PE=4 SV=1
MM1 pKa = 7.63LALDD5 pKa = 3.36QTGPYY10 pKa = 9.53LFQVRR15 pKa = 11.84HH16 pKa = 5.3GVALATGQDD25 pKa = 3.6DD26 pKa = 4.98VIDD29 pKa = 4.31GGAGNDD35 pKa = 3.82TLIGGPGNDD44 pKa = 4.03TIAGGSGWDD53 pKa = 3.33TAVYY57 pKa = 9.81SGNYY61 pKa = 9.49DD62 pKa = 3.35EE63 pKa = 5.01YY64 pKa = 11.25QITEE68 pKa = 4.24TDD70 pKa = 3.39TGWTVKK76 pKa = 10.6DD77 pKa = 3.8NRR79 pKa = 11.84QDD81 pKa = 3.71SPDD84 pKa = 3.62GTDD87 pKa = 3.4TISDD91 pKa = 3.57DD92 pKa = 3.98VEE94 pKa = 4.13NLEE97 pKa = 5.1FADD100 pKa = 3.67QTVSLVPTAEE110 pKa = 4.21EE111 pKa = 3.72IFIAEE116 pKa = 4.32GNDD119 pKa = 3.4GLRR122 pKa = 11.84NNEE125 pKa = 4.03AYY127 pKa = 7.96FTAGMMRR134 pKa = 11.84VMADD138 pKa = 3.31FMKK141 pKa = 10.64ASYY144 pKa = 10.61SLEE147 pKa = 3.77EE148 pKa = 4.18SEE150 pKa = 4.71NRR152 pKa = 11.84VINDD156 pKa = 3.38EE157 pKa = 4.19NGGADD162 pKa = 3.25TALTEE167 pKa = 5.51LYY169 pKa = 10.47DD170 pKa = 4.15QGWQPVFDD178 pKa = 5.33LIIPGVEE185 pKa = 3.51ATTILTVEE193 pKa = 4.95DD194 pKa = 3.77LTASYY199 pKa = 10.74QEE201 pKa = 4.11EE202 pKa = 4.22DD203 pKa = 3.66AEE205 pKa = 4.34QSGLASQMVSNGFSQEE221 pKa = 3.62TGLFTNGNAAALILRR236 pKa = 11.84SGDD239 pKa = 3.66SLVLSFRR246 pKa = 11.84GTNDD250 pKa = 2.89TGSPNDD256 pKa = 4.06DD257 pKa = 3.35SGILNTSDD265 pKa = 4.06PNNIIYY271 pKa = 9.46PDD273 pKa = 4.34RR274 pKa = 11.84IHH276 pKa = 6.87WGGAASIFDD285 pKa = 3.8SWSEE289 pKa = 3.94WEE291 pKa = 4.3GRR293 pKa = 11.84DD294 pKa = 4.13ANMSDD299 pKa = 3.97HH300 pKa = 6.48YY301 pKa = 11.84ALFAPLITAIEE312 pKa = 4.31NYY314 pKa = 9.89VGNADD319 pKa = 3.3NGINHH324 pKa = 6.12VYY326 pKa = 9.13ATGHH330 pKa = 5.61SLGGAMAIEE339 pKa = 4.68FTSQHH344 pKa = 6.03EE345 pKa = 4.38DD346 pKa = 3.2MVDD349 pKa = 3.3QTITFAAPAFTNEE362 pKa = 3.7NLSRR366 pKa = 11.84EE367 pKa = 4.16EE368 pKa = 5.79FEE370 pKa = 6.0DD371 pKa = 4.84LDD373 pKa = 3.88NLTQVEE379 pKa = 4.42ILFDD383 pKa = 4.47PVPMTWDD390 pKa = 2.96SLGGVNRR397 pKa = 11.84PGQLIRR403 pKa = 11.84LAGDD407 pKa = 3.61GTSTPIDD414 pKa = 3.58EE415 pKa = 4.85NFDD418 pKa = 4.19NILGADD424 pKa = 3.82SVNSNHH430 pKa = 5.17STNYY434 pKa = 9.76YY435 pKa = 9.03RR436 pKa = 11.84TITDD440 pKa = 4.92AIDD443 pKa = 3.52SVSWQAIINGTDD455 pKa = 3.06AQDD458 pKa = 3.14IYY460 pKa = 11.01LDD462 pKa = 3.54GMRR465 pKa = 11.84EE466 pKa = 3.93VEE468 pKa = 3.98NRR470 pKa = 11.84EE471 pKa = 3.65EE472 pKa = 4.32SFVVSVNDD480 pKa = 4.74DD481 pKa = 3.86VITYY485 pKa = 9.78DD486 pKa = 4.08PNWNDD491 pKa = 2.99FFDD494 pKa = 5.4GYY496 pKa = 11.06FGDD499 pKa = 4.04EE500 pKa = 4.47FGFDD504 pKa = 3.42AYY506 pKa = 11.38YY507 pKa = 10.89GGNGNDD513 pKa = 3.59HH514 pKa = 6.22LTGGEE519 pKa = 3.99EE520 pKa = 4.16GDD522 pKa = 3.62ILFGGGDD529 pKa = 3.79NDD531 pKa = 3.59RR532 pKa = 11.84LAGRR536 pKa = 11.84GGDD539 pKa = 3.72DD540 pKa = 2.82IFYY543 pKa = 10.58GGSGNDD549 pKa = 3.65RR550 pKa = 11.84IHH552 pKa = 7.29GGEE555 pKa = 4.03GTDD558 pKa = 3.29TVVYY562 pKa = 10.5ASSADD567 pKa = 3.57NYY569 pKa = 11.46DD570 pKa = 3.55FAIEE574 pKa = 3.86NWALTVKK581 pKa = 10.36PHH583 pKa = 6.64DD584 pKa = 3.98TNEE587 pKa = 3.76TDD589 pKa = 2.98TLYY592 pKa = 10.95SIEE595 pKa = 3.83QLGFGGEE602 pKa = 4.25QPVSVASEE610 pKa = 3.71LSYY613 pKa = 11.48TNNQLLHH620 pKa = 6.4MYY622 pKa = 10.74APILAMNADD631 pKa = 4.42DD632 pKa = 4.24YY633 pKa = 11.79VPTRR637 pKa = 11.84IEE639 pKa = 3.85AFLDD643 pKa = 3.35HH644 pKa = 7.44AILFDD649 pKa = 4.17VVWGNNPLAVGNNISGIVYY668 pKa = 10.33EE669 pKa = 5.19EE670 pKa = 4.15GVQYY674 pKa = 10.92NINTFVTGTSSIEE687 pKa = 3.88NAEE690 pKa = 4.14LVDD693 pKa = 4.5DD694 pKa = 4.4VSNLLSEE701 pKa = 4.66EE702 pKa = 4.15FHH704 pKa = 6.51SHH706 pKa = 8.01LIRR709 pKa = 11.84SNDD712 pKa = 3.25YY713 pKa = 10.3YY714 pKa = 11.64LDD716 pKa = 4.88FISGTEE722 pKa = 3.92NSATRR727 pKa = 11.84VDD729 pKa = 4.05PQTEE733 pKa = 4.33VPKK736 pKa = 10.8SYY738 pKa = 11.14GRR740 pKa = 11.84MDD742 pKa = 4.01TNPGSTGSGEE752 pKa = 3.93WADD755 pKa = 3.65SNEE758 pKa = 4.16NQFSFDD764 pKa = 3.38PTVIDD769 pKa = 3.88DD770 pKa = 3.61QYY772 pKa = 10.74GASVYY777 pKa = 10.57GRR779 pKa = 11.84VVTPEE784 pKa = 4.22AGDD787 pKa = 4.25AIYY790 pKa = 10.08LQYY793 pKa = 11.4YY794 pKa = 8.41FFYY797 pKa = 11.07LEE799 pKa = 4.18NHH801 pKa = 4.83WTSPISRR808 pKa = 11.84GFHH811 pKa = 4.81EE812 pKa = 5.25ADD814 pKa = 2.99WEE816 pKa = 4.39FMQIEE821 pKa = 4.47LDD823 pKa = 4.04KK824 pKa = 11.13DD825 pKa = 3.63TLLPDD830 pKa = 3.61TFMSSIHH837 pKa = 7.05LDD839 pKa = 3.19HH840 pKa = 6.74SQNKK844 pKa = 7.86PQYY847 pKa = 8.91PNQIHH852 pKa = 4.45QQSYY856 pKa = 9.43RR857 pKa = 11.84FQGLQSVYY865 pKa = 9.99HH866 pKa = 5.87YY867 pKa = 9.19PHH869 pKa = 6.8HH870 pKa = 6.8HH871 pKa = 5.81QLHH874 pKa = 6.29HH875 pKa = 6.68PVLL878 pKa = 4.79
MM1 pKa = 7.63LALDD5 pKa = 3.36QTGPYY10 pKa = 9.53LFQVRR15 pKa = 11.84HH16 pKa = 5.3GVALATGQDD25 pKa = 3.6DD26 pKa = 4.98VIDD29 pKa = 4.31GGAGNDD35 pKa = 3.82TLIGGPGNDD44 pKa = 4.03TIAGGSGWDD53 pKa = 3.33TAVYY57 pKa = 9.81SGNYY61 pKa = 9.49DD62 pKa = 3.35EE63 pKa = 5.01YY64 pKa = 11.25QITEE68 pKa = 4.24TDD70 pKa = 3.39TGWTVKK76 pKa = 10.6DD77 pKa = 3.8NRR79 pKa = 11.84QDD81 pKa = 3.71SPDD84 pKa = 3.62GTDD87 pKa = 3.4TISDD91 pKa = 3.57DD92 pKa = 3.98VEE94 pKa = 4.13NLEE97 pKa = 5.1FADD100 pKa = 3.67QTVSLVPTAEE110 pKa = 4.21EE111 pKa = 3.72IFIAEE116 pKa = 4.32GNDD119 pKa = 3.4GLRR122 pKa = 11.84NNEE125 pKa = 4.03AYY127 pKa = 7.96FTAGMMRR134 pKa = 11.84VMADD138 pKa = 3.31FMKK141 pKa = 10.64ASYY144 pKa = 10.61SLEE147 pKa = 3.77EE148 pKa = 4.18SEE150 pKa = 4.71NRR152 pKa = 11.84VINDD156 pKa = 3.38EE157 pKa = 4.19NGGADD162 pKa = 3.25TALTEE167 pKa = 5.51LYY169 pKa = 10.47DD170 pKa = 4.15QGWQPVFDD178 pKa = 5.33LIIPGVEE185 pKa = 3.51ATTILTVEE193 pKa = 4.95DD194 pKa = 3.77LTASYY199 pKa = 10.74QEE201 pKa = 4.11EE202 pKa = 4.22DD203 pKa = 3.66AEE205 pKa = 4.34QSGLASQMVSNGFSQEE221 pKa = 3.62TGLFTNGNAAALILRR236 pKa = 11.84SGDD239 pKa = 3.66SLVLSFRR246 pKa = 11.84GTNDD250 pKa = 2.89TGSPNDD256 pKa = 4.06DD257 pKa = 3.35SGILNTSDD265 pKa = 4.06PNNIIYY271 pKa = 9.46PDD273 pKa = 4.34RR274 pKa = 11.84IHH276 pKa = 6.87WGGAASIFDD285 pKa = 3.8SWSEE289 pKa = 3.94WEE291 pKa = 4.3GRR293 pKa = 11.84DD294 pKa = 4.13ANMSDD299 pKa = 3.97HH300 pKa = 6.48YY301 pKa = 11.84ALFAPLITAIEE312 pKa = 4.31NYY314 pKa = 9.89VGNADD319 pKa = 3.3NGINHH324 pKa = 6.12VYY326 pKa = 9.13ATGHH330 pKa = 5.61SLGGAMAIEE339 pKa = 4.68FTSQHH344 pKa = 6.03EE345 pKa = 4.38DD346 pKa = 3.2MVDD349 pKa = 3.3QTITFAAPAFTNEE362 pKa = 3.7NLSRR366 pKa = 11.84EE367 pKa = 4.16EE368 pKa = 5.79FEE370 pKa = 6.0DD371 pKa = 4.84LDD373 pKa = 3.88NLTQVEE379 pKa = 4.42ILFDD383 pKa = 4.47PVPMTWDD390 pKa = 2.96SLGGVNRR397 pKa = 11.84PGQLIRR403 pKa = 11.84LAGDD407 pKa = 3.61GTSTPIDD414 pKa = 3.58EE415 pKa = 4.85NFDD418 pKa = 4.19NILGADD424 pKa = 3.82SVNSNHH430 pKa = 5.17STNYY434 pKa = 9.76YY435 pKa = 9.03RR436 pKa = 11.84TITDD440 pKa = 4.92AIDD443 pKa = 3.52SVSWQAIINGTDD455 pKa = 3.06AQDD458 pKa = 3.14IYY460 pKa = 11.01LDD462 pKa = 3.54GMRR465 pKa = 11.84EE466 pKa = 3.93VEE468 pKa = 3.98NRR470 pKa = 11.84EE471 pKa = 3.65EE472 pKa = 4.32SFVVSVNDD480 pKa = 4.74DD481 pKa = 3.86VITYY485 pKa = 9.78DD486 pKa = 4.08PNWNDD491 pKa = 2.99FFDD494 pKa = 5.4GYY496 pKa = 11.06FGDD499 pKa = 4.04EE500 pKa = 4.47FGFDD504 pKa = 3.42AYY506 pKa = 11.38YY507 pKa = 10.89GGNGNDD513 pKa = 3.59HH514 pKa = 6.22LTGGEE519 pKa = 3.99EE520 pKa = 4.16GDD522 pKa = 3.62ILFGGGDD529 pKa = 3.79NDD531 pKa = 3.59RR532 pKa = 11.84LAGRR536 pKa = 11.84GGDD539 pKa = 3.72DD540 pKa = 2.82IFYY543 pKa = 10.58GGSGNDD549 pKa = 3.65RR550 pKa = 11.84IHH552 pKa = 7.29GGEE555 pKa = 4.03GTDD558 pKa = 3.29TVVYY562 pKa = 10.5ASSADD567 pKa = 3.57NYY569 pKa = 11.46DD570 pKa = 3.55FAIEE574 pKa = 3.86NWALTVKK581 pKa = 10.36PHH583 pKa = 6.64DD584 pKa = 3.98TNEE587 pKa = 3.76TDD589 pKa = 2.98TLYY592 pKa = 10.95SIEE595 pKa = 3.83QLGFGGEE602 pKa = 4.25QPVSVASEE610 pKa = 3.71LSYY613 pKa = 11.48TNNQLLHH620 pKa = 6.4MYY622 pKa = 10.74APILAMNADD631 pKa = 4.42DD632 pKa = 4.24YY633 pKa = 11.79VPTRR637 pKa = 11.84IEE639 pKa = 3.85AFLDD643 pKa = 3.35HH644 pKa = 7.44AILFDD649 pKa = 4.17VVWGNNPLAVGNNISGIVYY668 pKa = 10.33EE669 pKa = 5.19EE670 pKa = 4.15GVQYY674 pKa = 10.92NINTFVTGTSSIEE687 pKa = 3.88NAEE690 pKa = 4.14LVDD693 pKa = 4.5DD694 pKa = 4.4VSNLLSEE701 pKa = 4.66EE702 pKa = 4.15FHH704 pKa = 6.51SHH706 pKa = 8.01LIRR709 pKa = 11.84SNDD712 pKa = 3.25YY713 pKa = 10.3YY714 pKa = 11.64LDD716 pKa = 4.88FISGTEE722 pKa = 3.92NSATRR727 pKa = 11.84VDD729 pKa = 4.05PQTEE733 pKa = 4.33VPKK736 pKa = 10.8SYY738 pKa = 11.14GRR740 pKa = 11.84MDD742 pKa = 4.01TNPGSTGSGEE752 pKa = 3.93WADD755 pKa = 3.65SNEE758 pKa = 4.16NQFSFDD764 pKa = 3.38PTVIDD769 pKa = 3.88DD770 pKa = 3.61QYY772 pKa = 10.74GASVYY777 pKa = 10.57GRR779 pKa = 11.84VVTPEE784 pKa = 4.22AGDD787 pKa = 4.25AIYY790 pKa = 10.08LQYY793 pKa = 11.4YY794 pKa = 8.41FFYY797 pKa = 11.07LEE799 pKa = 4.18NHH801 pKa = 4.83WTSPISRR808 pKa = 11.84GFHH811 pKa = 4.81EE812 pKa = 5.25ADD814 pKa = 2.99WEE816 pKa = 4.39FMQIEE821 pKa = 4.47LDD823 pKa = 4.04KK824 pKa = 11.13DD825 pKa = 3.63TLLPDD830 pKa = 3.61TFMSSIHH837 pKa = 7.05LDD839 pKa = 3.19HH840 pKa = 6.74SQNKK844 pKa = 7.86PQYY847 pKa = 8.91PNQIHH852 pKa = 4.45QQSYY856 pKa = 9.43RR857 pKa = 11.84FQGLQSVYY865 pKa = 9.99HH866 pKa = 5.87YY867 pKa = 9.19PHH869 pKa = 6.8HH870 pKa = 6.8HH871 pKa = 5.81QLHH874 pKa = 6.29HH875 pKa = 6.68PVLL878 pKa = 4.79
Molecular weight: 96.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M1JJG4|A0A0M1JJG4_9GAMM Uncharacterized protein OS=Achromatium sp. WMS3 OX=1604836 GN=TI05_01120 PE=4 SV=1
MM1 pKa = 7.32KK2 pKa = 10.12RR3 pKa = 11.84YY4 pKa = 9.47IHH6 pKa = 6.89LLVAWLFIITAQQTYY21 pKa = 9.6AADD24 pKa = 3.62VGGSIPNTTEE34 pKa = 3.17ATHH37 pKa = 6.75AVTTRR42 pKa = 11.84KK43 pKa = 9.3IVLDD47 pKa = 3.64PCIKK51 pKa = 9.01TRR53 pKa = 11.84KK54 pKa = 9.04IVFNGSKK61 pKa = 9.05TPCRR65 pKa = 11.84RR66 pKa = 11.84YY67 pKa = 10.11KK68 pKa = 10.49RR69 pKa = 11.84PKK71 pKa = 7.74TSSRR75 pKa = 11.84PSCYY79 pKa = 10.13ARR81 pKa = 11.84GPRR84 pKa = 11.84TRR86 pKa = 11.84KK87 pKa = 9.38IVWGKK92 pKa = 8.89EE93 pKa = 3.65YY94 pKa = 10.92CRR96 pKa = 4.24
MM1 pKa = 7.32KK2 pKa = 10.12RR3 pKa = 11.84YY4 pKa = 9.47IHH6 pKa = 6.89LLVAWLFIITAQQTYY21 pKa = 9.6AADD24 pKa = 3.62VGGSIPNTTEE34 pKa = 3.17ATHH37 pKa = 6.75AVTTRR42 pKa = 11.84KK43 pKa = 9.3IVLDD47 pKa = 3.64PCIKK51 pKa = 9.01TRR53 pKa = 11.84KK54 pKa = 9.04IVFNGSKK61 pKa = 9.05TPCRR65 pKa = 11.84RR66 pKa = 11.84YY67 pKa = 10.11KK68 pKa = 10.49RR69 pKa = 11.84PKK71 pKa = 7.74TSSRR75 pKa = 11.84PSCYY79 pKa = 10.13ARR81 pKa = 11.84GPRR84 pKa = 11.84TRR86 pKa = 11.84KK87 pKa = 9.38IVWGKK92 pKa = 8.89EE93 pKa = 3.65YY94 pKa = 10.92CRR96 pKa = 4.24
Molecular weight: 11.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
775224 |
26 |
3350 |
250.2 |
28.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.061 ± 0.054 | 1.075 ± 0.022 |
5.139 ± 0.053 | 5.471 ± 0.046 |
3.904 ± 0.036 | 6.194 ± 0.048 |
2.395 ± 0.024 | 7.909 ± 0.045 |
5.584 ± 0.047 | 10.687 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.024 | 4.863 ± 0.042 |
4.377 ± 0.032 | 4.994 ± 0.046 |
4.887 ± 0.042 | 5.65 ± 0.04 |
6.181 ± 0.06 | 5.8 ± 0.043 |
1.359 ± 0.02 | 3.324 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
