
Tannerella sp. oral taxon 808
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Tannerellaceae; Tannerella; unclassified Tannerella
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
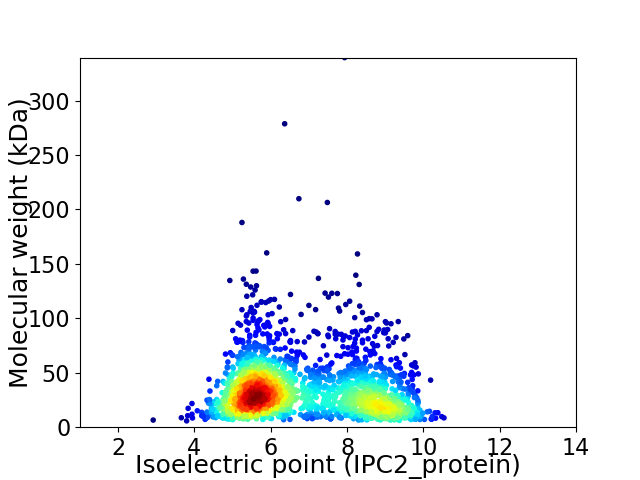
Virtual 2D-PAGE plot for 1968 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N8NL91|A0A2N8NL91_9BACT 30S ribosomal protein S18 OS=Tannerella sp. oral taxon 808 OX=712711 GN=rpsR PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.13YY4 pKa = 10.49RR5 pKa = 11.84CTVCDD10 pKa = 3.77YY11 pKa = 10.76IYY13 pKa = 10.64DD14 pKa = 4.11PEE16 pKa = 5.15VGDD19 pKa = 4.16PDD21 pKa = 4.6GGIDD25 pKa = 3.47PGTAFEE31 pKa = 5.39EE32 pKa = 4.74IPDD35 pKa = 3.73DD36 pKa = 4.16WVCPLCGVGKK46 pKa = 10.53EE47 pKa = 4.22DD48 pKa = 4.73FEE50 pKa = 4.75PVV52 pKa = 2.91
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.13YY4 pKa = 10.49RR5 pKa = 11.84CTVCDD10 pKa = 3.77YY11 pKa = 10.76IYY13 pKa = 10.64DD14 pKa = 4.11PEE16 pKa = 5.15VGDD19 pKa = 4.16PDD21 pKa = 4.6GGIDD25 pKa = 3.47PGTAFEE31 pKa = 5.39EE32 pKa = 4.74IPDD35 pKa = 3.73DD36 pKa = 4.16WVCPLCGVGKK46 pKa = 10.53EE47 pKa = 4.22DD48 pKa = 4.73FEE50 pKa = 4.75PVV52 pKa = 2.91
Molecular weight: 5.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N8N8Z9|A0A2N8N8Z9_9BACT DZANK-type domain-containing protein OS=Tannerella sp. oral taxon 808 OX=712711 GN=BHU16_02795 PE=4 SV=1
MM1 pKa = 6.96TTTLLTTLAVLFGILMLVLTLRR23 pKa = 11.84EE24 pKa = 4.01KK25 pKa = 10.16QRR27 pKa = 11.84EE28 pKa = 3.96RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 10.54LSGSAPDD38 pKa = 3.28APYY41 pKa = 10.22RR42 pKa = 11.84QQPPPVNPPSTDD54 pKa = 3.45RR55 pKa = 11.84DD56 pKa = 3.95PLNDD60 pKa = 3.5IYY62 pKa = 11.32GADD65 pKa = 3.9AIARR69 pKa = 11.84CAAQLCTTEE78 pKa = 4.59GEE80 pKa = 4.26LRR82 pKa = 11.84DD83 pKa = 3.8VLRR86 pKa = 11.84DD87 pKa = 3.62IRR89 pKa = 11.84WAGYY93 pKa = 4.62TTFWMAKK100 pKa = 9.09RR101 pKa = 11.84RR102 pKa = 11.84GGYY105 pKa = 10.01RR106 pKa = 11.84NIEE109 pKa = 4.3APTPPLKK116 pKa = 10.62AIQRR120 pKa = 11.84AILDD124 pKa = 3.79RR125 pKa = 11.84LLTPLPVHH133 pKa = 6.18TAATGFRR140 pKa = 11.84PGLSIADD147 pKa = 3.63NARR150 pKa = 11.84PHH152 pKa = 6.7LGQRR156 pKa = 11.84RR157 pKa = 11.84ALKK160 pKa = 9.8TDD162 pKa = 2.9LHH164 pKa = 7.06DD165 pKa = 3.87FFGSIRR171 pKa = 11.84RR172 pKa = 11.84PAIRR176 pKa = 11.84RR177 pKa = 11.84VFRR180 pKa = 11.84DD181 pKa = 3.17MGFDD185 pKa = 3.27AHH187 pKa = 5.7ITKK190 pKa = 10.26VLVDD194 pKa = 4.12LCTHH198 pKa = 7.01RR199 pKa = 11.84NRR201 pKa = 11.84LPQGAPTSPALSNLVAAKK219 pKa = 9.52MDD221 pKa = 3.59EE222 pKa = 4.12RR223 pKa = 11.84LTALSLRR230 pKa = 11.84RR231 pKa = 11.84GLTYY235 pKa = 10.15TRR237 pKa = 11.84YY238 pKa = 10.52ADD240 pKa = 4.44DD241 pKa = 4.12LTFSGDD247 pKa = 3.21SFNRR251 pKa = 11.84NEE253 pKa = 4.48LIADD257 pKa = 3.89IKK259 pKa = 11.03HH260 pKa = 5.67IATYY264 pKa = 10.93DD265 pKa = 3.61GFTLNAKK272 pKa = 7.98KK273 pKa = 8.7TRR275 pKa = 11.84LMASSRR281 pKa = 11.84RR282 pKa = 11.84IVTGLSIGSGEE293 pKa = 4.51KK294 pKa = 9.27LTLPRR299 pKa = 11.84EE300 pKa = 4.27RR301 pKa = 11.84KK302 pKa = 9.17RR303 pKa = 11.84EE304 pKa = 3.69IRR306 pKa = 11.84KK307 pKa = 8.87NVHH310 pKa = 5.95FVLKK314 pKa = 10.83YY315 pKa = 9.78GVTEE319 pKa = 3.61HH320 pKa = 7.31RR321 pKa = 11.84EE322 pKa = 4.27RR323 pKa = 11.84IHH325 pKa = 8.12SSDD328 pKa = 3.07PAYY331 pKa = 10.57VKK333 pKa = 10.62RR334 pKa = 11.84LIGEE338 pKa = 4.01LCFWRR343 pKa = 11.84SIEE346 pKa = 4.07PEE348 pKa = 3.93HH349 pKa = 6.68PFVNRR354 pKa = 11.84ALRR357 pKa = 11.84EE358 pKa = 3.99LRR360 pKa = 11.84ALRR363 pKa = 4.05
MM1 pKa = 6.96TTTLLTTLAVLFGILMLVLTLRR23 pKa = 11.84EE24 pKa = 4.01KK25 pKa = 10.16QRR27 pKa = 11.84EE28 pKa = 3.96RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 10.54LSGSAPDD38 pKa = 3.28APYY41 pKa = 10.22RR42 pKa = 11.84QQPPPVNPPSTDD54 pKa = 3.45RR55 pKa = 11.84DD56 pKa = 3.95PLNDD60 pKa = 3.5IYY62 pKa = 11.32GADD65 pKa = 3.9AIARR69 pKa = 11.84CAAQLCTTEE78 pKa = 4.59GEE80 pKa = 4.26LRR82 pKa = 11.84DD83 pKa = 3.8VLRR86 pKa = 11.84DD87 pKa = 3.62IRR89 pKa = 11.84WAGYY93 pKa = 4.62TTFWMAKK100 pKa = 9.09RR101 pKa = 11.84RR102 pKa = 11.84GGYY105 pKa = 10.01RR106 pKa = 11.84NIEE109 pKa = 4.3APTPPLKK116 pKa = 10.62AIQRR120 pKa = 11.84AILDD124 pKa = 3.79RR125 pKa = 11.84LLTPLPVHH133 pKa = 6.18TAATGFRR140 pKa = 11.84PGLSIADD147 pKa = 3.63NARR150 pKa = 11.84PHH152 pKa = 6.7LGQRR156 pKa = 11.84RR157 pKa = 11.84ALKK160 pKa = 9.8TDD162 pKa = 2.9LHH164 pKa = 7.06DD165 pKa = 3.87FFGSIRR171 pKa = 11.84RR172 pKa = 11.84PAIRR176 pKa = 11.84RR177 pKa = 11.84VFRR180 pKa = 11.84DD181 pKa = 3.17MGFDD185 pKa = 3.27AHH187 pKa = 5.7ITKK190 pKa = 10.26VLVDD194 pKa = 4.12LCTHH198 pKa = 7.01RR199 pKa = 11.84NRR201 pKa = 11.84LPQGAPTSPALSNLVAAKK219 pKa = 9.52MDD221 pKa = 3.59EE222 pKa = 4.12RR223 pKa = 11.84LTALSLRR230 pKa = 11.84RR231 pKa = 11.84GLTYY235 pKa = 10.15TRR237 pKa = 11.84YY238 pKa = 10.52ADD240 pKa = 4.44DD241 pKa = 4.12LTFSGDD247 pKa = 3.21SFNRR251 pKa = 11.84NEE253 pKa = 4.48LIADD257 pKa = 3.89IKK259 pKa = 11.03HH260 pKa = 5.67IATYY264 pKa = 10.93DD265 pKa = 3.61GFTLNAKK272 pKa = 7.98KK273 pKa = 8.7TRR275 pKa = 11.84LMASSRR281 pKa = 11.84RR282 pKa = 11.84IVTGLSIGSGEE293 pKa = 4.51KK294 pKa = 9.27LTLPRR299 pKa = 11.84EE300 pKa = 4.27RR301 pKa = 11.84KK302 pKa = 9.17RR303 pKa = 11.84EE304 pKa = 3.69IRR306 pKa = 11.84KK307 pKa = 8.87NVHH310 pKa = 5.95FVLKK314 pKa = 10.83YY315 pKa = 9.78GVTEE319 pKa = 3.61HH320 pKa = 7.31RR321 pKa = 11.84EE322 pKa = 4.27RR323 pKa = 11.84IHH325 pKa = 8.12SSDD328 pKa = 3.07PAYY331 pKa = 10.57VKK333 pKa = 10.62RR334 pKa = 11.84LIGEE338 pKa = 4.01LCFWRR343 pKa = 11.84SIEE346 pKa = 4.07PEE348 pKa = 3.93HH349 pKa = 6.68PFVNRR354 pKa = 11.84ALRR357 pKa = 11.84EE358 pKa = 3.99LRR360 pKa = 11.84ALRR363 pKa = 4.05
Molecular weight: 41.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
648991 |
52 |
3042 |
329.8 |
36.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.963 ± 0.062 | 1.114 ± 0.02 |
5.808 ± 0.038 | 6.184 ± 0.054 |
4.207 ± 0.033 | 7.039 ± 0.044 |
2.224 ± 0.025 | 6.083 ± 0.049 |
4.859 ± 0.057 | 9.481 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.027 | 3.739 ± 0.042 |
4.381 ± 0.032 | 3.086 ± 0.032 |
7.005 ± 0.052 | 5.513 ± 0.039 |
6.023 ± 0.051 | 6.644 ± 0.048 |
1.079 ± 0.019 | 3.9 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
