
Rotavirus A pheasant-tc/GER/10V0112H5/2010/G23P[37]
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus A; Rotavirus A isolates
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
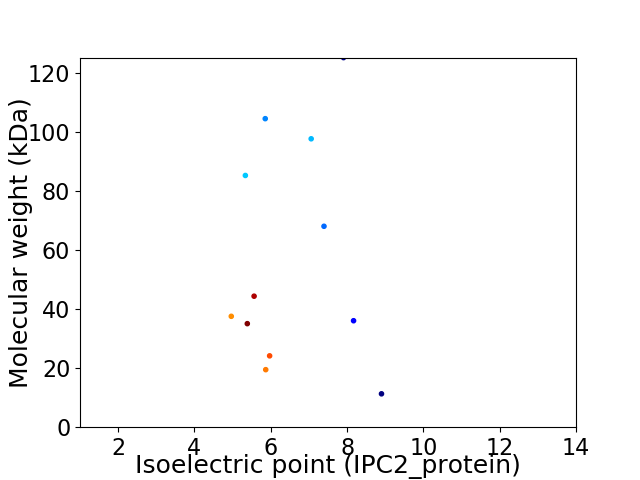
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
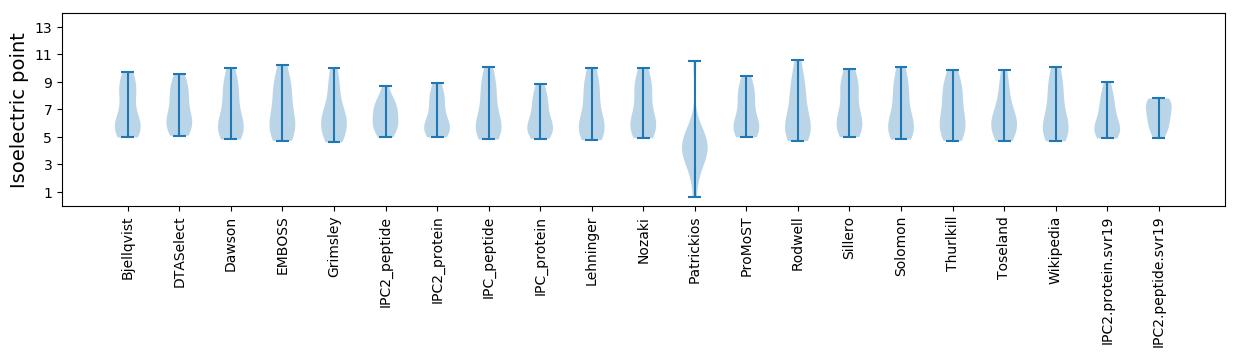
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0CSN4|L0CSN4_9REOV Non-structural protein 5 OS=Rotavirus A pheasant-tc/GER/10V0112H5/2010/G23P[37] OX=1271534 PE=4 SV=1
MM1 pKa = 7.49YY2 pKa = 9.41STEE5 pKa = 4.05CTSFFLEE12 pKa = 4.15IIFYY16 pKa = 10.44VILYY20 pKa = 7.18TIISSRR26 pKa = 11.84LSRR29 pKa = 11.84MSRR32 pKa = 11.84LFYY35 pKa = 10.16WIIIFIIFSSLCVSKK50 pKa = 10.92CSSQNFGINVPITGSLDD67 pKa = 3.41VTVLNSTQTQIGLTSTLCIYY87 pKa = 10.0YY88 pKa = 9.69PKK90 pKa = 10.12EE91 pKa = 3.83ASDD94 pKa = 4.14EE95 pKa = 4.34IKK97 pKa = 10.83DD98 pKa = 4.09SEE100 pKa = 4.32WKK102 pKa = 8.72NTVSQLLLTKK112 pKa = 9.96GWPTTSVYY120 pKa = 10.89LNEE123 pKa = 4.26YY124 pKa = 9.77NDD126 pKa = 4.06LAAFSNDD133 pKa = 2.55PKK135 pKa = 10.92LYY137 pKa = 9.75CDD139 pKa = 3.72YY140 pKa = 11.28NIVLVKK146 pKa = 9.17YY147 pKa = 8.33TGSNVALDD155 pKa = 3.47ISEE158 pKa = 4.28LAEE161 pKa = 3.99LLLYY165 pKa = 10.18EE166 pKa = 4.42WLCNPMDD173 pKa = 3.53VTLYY177 pKa = 10.62YY178 pKa = 9.99YY179 pKa = 10.53QQTSEE184 pKa = 4.23SNKK187 pKa = 8.2WIAMGTDD194 pKa = 3.77CTIKK198 pKa = 10.33VCPLNVQTLGIGCQTTDD215 pKa = 2.68VNTFEE220 pKa = 4.48QVTASEE226 pKa = 4.0QLAIIDD232 pKa = 3.76VVDD235 pKa = 4.36GVNHH239 pKa = 7.27KK240 pKa = 10.36INYY243 pKa = 7.41TLTTCTIKK251 pKa = 10.89NCIRR255 pKa = 11.84LNPRR259 pKa = 11.84GNVAIIQIGGPEE271 pKa = 3.96ILDD274 pKa = 3.32ISEE277 pKa = 4.74DD278 pKa = 3.66PMVVPKK284 pKa = 9.3MVRR287 pKa = 11.84MTRR290 pKa = 11.84INWKK294 pKa = 9.41KK295 pKa = 7.27WWQVFYY301 pKa = 10.64TIVDD305 pKa = 3.85YY306 pKa = 11.35INTIVNAMSKK316 pKa = 10.25RR317 pKa = 11.84ATSLDD322 pKa = 2.99ASSYY326 pKa = 7.88YY327 pKa = 10.84LRR329 pKa = 11.84VV330 pKa = 3.27
MM1 pKa = 7.49YY2 pKa = 9.41STEE5 pKa = 4.05CTSFFLEE12 pKa = 4.15IIFYY16 pKa = 10.44VILYY20 pKa = 7.18TIISSRR26 pKa = 11.84LSRR29 pKa = 11.84MSRR32 pKa = 11.84LFYY35 pKa = 10.16WIIIFIIFSSLCVSKK50 pKa = 10.92CSSQNFGINVPITGSLDD67 pKa = 3.41VTVLNSTQTQIGLTSTLCIYY87 pKa = 10.0YY88 pKa = 9.69PKK90 pKa = 10.12EE91 pKa = 3.83ASDD94 pKa = 4.14EE95 pKa = 4.34IKK97 pKa = 10.83DD98 pKa = 4.09SEE100 pKa = 4.32WKK102 pKa = 8.72NTVSQLLLTKK112 pKa = 9.96GWPTTSVYY120 pKa = 10.89LNEE123 pKa = 4.26YY124 pKa = 9.77NDD126 pKa = 4.06LAAFSNDD133 pKa = 2.55PKK135 pKa = 10.92LYY137 pKa = 9.75CDD139 pKa = 3.72YY140 pKa = 11.28NIVLVKK146 pKa = 9.17YY147 pKa = 8.33TGSNVALDD155 pKa = 3.47ISEE158 pKa = 4.28LAEE161 pKa = 3.99LLLYY165 pKa = 10.18EE166 pKa = 4.42WLCNPMDD173 pKa = 3.53VTLYY177 pKa = 10.62YY178 pKa = 9.99YY179 pKa = 10.53QQTSEE184 pKa = 4.23SNKK187 pKa = 8.2WIAMGTDD194 pKa = 3.77CTIKK198 pKa = 10.33VCPLNVQTLGIGCQTTDD215 pKa = 2.68VNTFEE220 pKa = 4.48QVTASEE226 pKa = 4.0QLAIIDD232 pKa = 3.76VVDD235 pKa = 4.36GVNHH239 pKa = 7.27KK240 pKa = 10.36INYY243 pKa = 7.41TLTTCTIKK251 pKa = 10.89NCIRR255 pKa = 11.84LNPRR259 pKa = 11.84GNVAIIQIGGPEE271 pKa = 3.96ILDD274 pKa = 3.32ISEE277 pKa = 4.74DD278 pKa = 3.66PMVVPKK284 pKa = 9.3MVRR287 pKa = 11.84MTRR290 pKa = 11.84INWKK294 pKa = 9.41KK295 pKa = 7.27WWQVFYY301 pKa = 10.64TIVDD305 pKa = 3.85YY306 pKa = 11.35INTIVNAMSKK316 pKa = 10.25RR317 pKa = 11.84ATSLDD322 pKa = 2.99ASSYY326 pKa = 7.88YY327 pKa = 10.84LRR329 pKa = 11.84VV330 pKa = 3.27
Molecular weight: 37.58 kDa
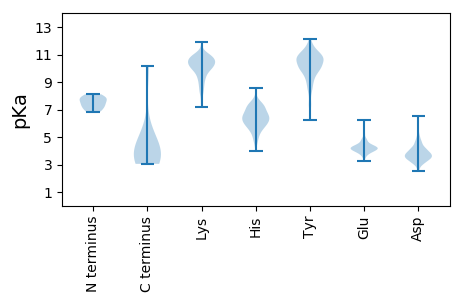
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0CRY4|L0CRY4_9REOV Intermediate capsid protein VP6 OS=Rotavirus A pheasant-tc/GER/10V0112H5/2010/G23P[37] OX=1271534 PE=3 SV=1
MM1 pKa = 6.85NHH3 pKa = 5.71LVKK6 pKa = 10.67RR7 pKa = 11.84QVLQGNLLVGVSSMYY22 pKa = 10.41RR23 pKa = 11.84CPKK26 pKa = 9.39KK27 pKa = 10.15HH28 pKa = 6.4LANTCLIGIQKK39 pKa = 8.46TLDD42 pKa = 3.36HH43 pKa = 7.23LIQHH47 pKa = 6.18QMTCPPNLLLNQMQLNQMLMLACRR71 pKa = 11.84WMLDD75 pKa = 3.03QWIVRR80 pKa = 11.84QEE82 pKa = 3.96TMQKK86 pKa = 8.57MKK88 pKa = 10.79SIFHH92 pKa = 5.94SLKK95 pKa = 10.27EE96 pKa = 3.95
MM1 pKa = 6.85NHH3 pKa = 5.71LVKK6 pKa = 10.67RR7 pKa = 11.84QVLQGNLLVGVSSMYY22 pKa = 10.41RR23 pKa = 11.84CPKK26 pKa = 9.39KK27 pKa = 10.15HH28 pKa = 6.4LANTCLIGIQKK39 pKa = 8.46TLDD42 pKa = 3.36HH43 pKa = 7.23LIQHH47 pKa = 6.18QMTCPPNLLLNQMQLNQMLMLACRR71 pKa = 11.84WMLDD75 pKa = 3.03QWIVRR80 pKa = 11.84QEE82 pKa = 3.96TMQKK86 pKa = 8.57MKK88 pKa = 10.79SIFHH92 pKa = 5.94SLKK95 pKa = 10.27EE96 pKa = 3.95
Molecular weight: 11.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5986 |
96 |
1089 |
498.8 |
57.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.396 ± 0.352 | 1.286 ± 0.368 |
6.215 ± 0.231 | 5.379 ± 0.269 |
4.227 ± 0.199 | 3.375 ± 0.343 |
1.57 ± 0.356 | 7.685 ± 0.271 |
6.799 ± 0.525 | 9.422 ± 0.323 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.74 ± 0.318 | 6.866 ± 0.339 |
3.291 ± 0.316 | 4.059 ± 0.531 |
4.744 ± 0.319 | 7.785 ± 0.779 |
6.498 ± 0.388 | 6.365 ± 0.223 |
1.169 ± 0.172 | 5.129 ± 0.609 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
