
Celeribacter marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Celeribacter
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
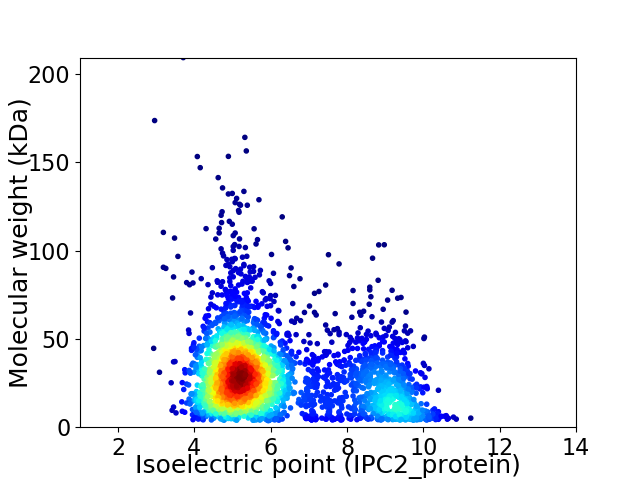
Virtual 2D-PAGE plot for 3134 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0A1A4|A0A0P0A1A4_9RHOB Uncharacterized protein OS=Celeribacter marinus OX=1397108 GN=IMCC12053_16 PE=4 SV=1
MM1 pKa = 7.24ATYY4 pKa = 10.3ILKK7 pKa = 10.63GYY9 pKa = 9.25AASSLSSTSGNLTAQVGHH27 pKa = 6.04VWRR30 pKa = 11.84LDD32 pKa = 3.78PSWDD36 pKa = 3.44NSNDD40 pKa = 3.1HH41 pKa = 6.28LTYY44 pKa = 10.38TINDD48 pKa = 3.61DD49 pKa = 3.77DD50 pKa = 4.14VRR52 pKa = 11.84FGGDD56 pKa = 2.97ATINEE61 pKa = 4.36VGDD64 pKa = 5.1DD65 pKa = 3.85FTQTAVVTNVSGGTVASGGVYY86 pKa = 10.65LEE88 pKa = 4.37DD89 pKa = 4.83QITLSDD95 pKa = 3.73GQGNLFDD102 pKa = 4.23VYY104 pKa = 10.79SVEE107 pKa = 4.21IGGTQVGVVSNPEE120 pKa = 3.94LVPGVTYY127 pKa = 10.37QVSSVSNVSAFNLPDD142 pKa = 3.87YY143 pKa = 11.47VDD145 pKa = 4.0MATPTYY151 pKa = 10.86DD152 pKa = 3.66PDD154 pKa = 4.22LAQTYY159 pKa = 10.85NGGAYY164 pKa = 9.96NDD166 pKa = 4.44EE167 pKa = 4.26IVAGAGDD174 pKa = 4.55DD175 pKa = 4.37IINAGAGADD184 pKa = 3.99SIEE187 pKa = 4.41GGAGNDD193 pKa = 3.52TIYY196 pKa = 10.9YY197 pKa = 6.62GTGGATQADD206 pKa = 4.5GDD208 pKa = 4.36TVYY211 pKa = 11.07GGDD214 pKa = 4.1GDD216 pKa = 6.2DD217 pKa = 5.14IIDD220 pKa = 4.32DD221 pKa = 3.93EE222 pKa = 5.68AVVQHH227 pKa = 6.56NYY229 pKa = 10.99DD230 pKa = 3.37DD231 pKa = 3.96TLYY234 pKa = 11.16GGAGNDD240 pKa = 3.5TIYY243 pKa = 11.09AGGGDD248 pKa = 3.83DD249 pKa = 4.97EE250 pKa = 5.63VYY252 pKa = 11.35GDD254 pKa = 5.47ADD256 pKa = 3.68NDD258 pKa = 4.39WIHH261 pKa = 6.97GEE263 pKa = 4.11GGHH266 pKa = 7.17DD267 pKa = 3.76EE268 pKa = 4.29LHH270 pKa = 6.72GGTGDD275 pKa = 3.46DD276 pKa = 4.15TIYY279 pKa = 11.24GGDD282 pKa = 4.04GYY284 pKa = 11.24DD285 pKa = 5.61LIWGDD290 pKa = 4.65DD291 pKa = 4.11GNDD294 pKa = 3.31TLYY297 pKa = 11.27GGNDD301 pKa = 3.76DD302 pKa = 3.7DD303 pKa = 4.98TIYY306 pKa = 11.07GGAGIDD312 pKa = 3.75VLYY315 pKa = 11.07GDD317 pKa = 5.3AGNDD321 pKa = 3.73TLSGDD326 pKa = 3.35AGGDD330 pKa = 3.36HH331 pKa = 7.45LYY333 pKa = 11.15GGTGNDD339 pKa = 3.42TMSGGTGNDD348 pKa = 2.65SFYY351 pKa = 11.46VLEE354 pKa = 4.81GDD356 pKa = 4.3GTSTMYY362 pKa = 11.24GDD364 pKa = 5.56DD365 pKa = 4.0DD366 pKa = 4.15WDD368 pKa = 4.08TVDD371 pKa = 4.65FAYY374 pKa = 9.53STGSAVDD381 pKa = 3.43VTYY384 pKa = 11.18SGDD387 pKa = 3.41NAFNFAFDD395 pKa = 3.96TGNSSGSGNSMEE407 pKa = 4.68NFILTGQNDD416 pKa = 4.05TVDD419 pKa = 3.63ASLDD423 pKa = 3.73ATGATYY429 pKa = 10.66DD430 pKa = 4.44LRR432 pKa = 11.84AGDD435 pKa = 3.89DD436 pKa = 3.71TFTGGSGNDD445 pKa = 3.14AVYY448 pKa = 10.61GGDD451 pKa = 3.63GADD454 pKa = 4.08TITLGAGGDD463 pKa = 3.93YY464 pKa = 11.39AEE466 pKa = 5.31GGAGNDD472 pKa = 4.05TIYY475 pKa = 11.44GDD477 pKa = 4.85AGEE480 pKa = 4.48DD481 pKa = 3.51WIEE484 pKa = 4.24GGTGNDD490 pKa = 3.32TLYY493 pKa = 11.28GGDD496 pKa = 3.73DD497 pKa = 3.94ADD499 pKa = 3.72TFFYY503 pKa = 10.99SDD505 pKa = 2.85GWGSDD510 pKa = 3.4TVFGGAGISDD520 pKa = 4.57ADD522 pKa = 3.78TLDD525 pKa = 3.7FSNVTTDD532 pKa = 3.25VTVTFSDD539 pKa = 3.89WEE541 pKa = 4.71DD542 pKa = 3.39GTATSGADD550 pKa = 2.98SVNFDD555 pKa = 3.95NIEE558 pKa = 4.4AIRR561 pKa = 11.84GGSGADD567 pKa = 3.73TIDD570 pKa = 3.79ASPDD574 pKa = 3.31GSGLLLDD581 pKa = 5.1GGDD584 pKa = 4.12GGDD587 pKa = 3.91TIIGGSGDD595 pKa = 3.73DD596 pKa = 3.85EE597 pKa = 5.88LYY599 pKa = 11.23GGDD602 pKa = 4.42GVDD605 pKa = 3.68TLFGGAGNDD614 pKa = 3.98AIDD617 pKa = 4.19GGRR620 pKa = 11.84GDD622 pKa = 3.86DD623 pKa = 4.44TITGGAGDD631 pKa = 4.12DD632 pKa = 4.62TITSSNGQDD641 pKa = 2.75TMVFADD647 pKa = 4.44GFGNDD652 pKa = 4.39IITDD656 pKa = 4.3FDD658 pKa = 4.07FTDD661 pKa = 4.05SNSDD665 pKa = 3.35GYY667 pKa = 9.63TDD669 pKa = 4.16DD670 pKa = 3.94QLDD673 pKa = 3.65VSALTDD679 pKa = 3.47SDD681 pKa = 4.3GNPVNAWDD689 pKa = 4.33VVVSDD694 pKa = 5.17DD695 pKa = 4.01GLGNAVLTFPNGEE708 pKa = 4.57SITLTGIAPTQVDD721 pKa = 4.55GAQQMHH727 pKa = 7.52AIGIPCFVSGTRR739 pKa = 11.84IMTPSGNRR747 pKa = 11.84LIEE750 pKa = 4.09EE751 pKa = 4.4LRR753 pKa = 11.84HH754 pKa = 5.69GDD756 pKa = 4.05LVSTLNSGAQPILWAGRR773 pKa = 11.84TTLKK777 pKa = 10.35RR778 pKa = 11.84ARR780 pKa = 11.84LALQPDD786 pKa = 4.75LLPIRR791 pKa = 11.84IRR793 pKa = 11.84DD794 pKa = 3.75GTLGNSGDD802 pKa = 3.7LWVSPQHH809 pKa = 6.59GLVLPDD815 pKa = 4.03LCSHH819 pKa = 6.81AKK821 pKa = 10.45AGVLVRR827 pKa = 11.84AKK829 pKa = 10.41HH830 pKa = 5.46LLEE833 pKa = 5.93HH834 pKa = 6.59GDD836 pKa = 3.36GRR838 pKa = 11.84VRR840 pKa = 11.84RR841 pKa = 11.84AKK843 pKa = 10.23GSRR846 pKa = 11.84DD847 pKa = 3.25VTYY850 pKa = 10.73HH851 pKa = 6.4HH852 pKa = 7.25LLLPRR857 pKa = 11.84HH858 pKa = 5.86ALILANGCASEE869 pKa = 4.26SLFPGRR875 pKa = 11.84FALSGFDD882 pKa = 3.19RR883 pKa = 11.84AAKK886 pKa = 10.62AEE888 pKa = 4.1LFDD891 pKa = 5.35LFPALEE897 pKa = 4.51TILRR901 pKa = 11.84SGAPATDD908 pKa = 3.61AARR911 pKa = 11.84MYY913 pKa = 8.49GQPVLRR919 pKa = 11.84FAKK922 pKa = 10.19GRR924 pKa = 11.84DD925 pKa = 2.85WRR927 pKa = 11.84AGHH930 pKa = 6.19IRR932 pKa = 11.84LPKK935 pKa = 9.96RR936 pKa = 11.84AA937 pKa = 3.85
MM1 pKa = 7.24ATYY4 pKa = 10.3ILKK7 pKa = 10.63GYY9 pKa = 9.25AASSLSSTSGNLTAQVGHH27 pKa = 6.04VWRR30 pKa = 11.84LDD32 pKa = 3.78PSWDD36 pKa = 3.44NSNDD40 pKa = 3.1HH41 pKa = 6.28LTYY44 pKa = 10.38TINDD48 pKa = 3.61DD49 pKa = 3.77DD50 pKa = 4.14VRR52 pKa = 11.84FGGDD56 pKa = 2.97ATINEE61 pKa = 4.36VGDD64 pKa = 5.1DD65 pKa = 3.85FTQTAVVTNVSGGTVASGGVYY86 pKa = 10.65LEE88 pKa = 4.37DD89 pKa = 4.83QITLSDD95 pKa = 3.73GQGNLFDD102 pKa = 4.23VYY104 pKa = 10.79SVEE107 pKa = 4.21IGGTQVGVVSNPEE120 pKa = 3.94LVPGVTYY127 pKa = 10.37QVSSVSNVSAFNLPDD142 pKa = 3.87YY143 pKa = 11.47VDD145 pKa = 4.0MATPTYY151 pKa = 10.86DD152 pKa = 3.66PDD154 pKa = 4.22LAQTYY159 pKa = 10.85NGGAYY164 pKa = 9.96NDD166 pKa = 4.44EE167 pKa = 4.26IVAGAGDD174 pKa = 4.55DD175 pKa = 4.37IINAGAGADD184 pKa = 3.99SIEE187 pKa = 4.41GGAGNDD193 pKa = 3.52TIYY196 pKa = 10.9YY197 pKa = 6.62GTGGATQADD206 pKa = 4.5GDD208 pKa = 4.36TVYY211 pKa = 11.07GGDD214 pKa = 4.1GDD216 pKa = 6.2DD217 pKa = 5.14IIDD220 pKa = 4.32DD221 pKa = 3.93EE222 pKa = 5.68AVVQHH227 pKa = 6.56NYY229 pKa = 10.99DD230 pKa = 3.37DD231 pKa = 3.96TLYY234 pKa = 11.16GGAGNDD240 pKa = 3.5TIYY243 pKa = 11.09AGGGDD248 pKa = 3.83DD249 pKa = 4.97EE250 pKa = 5.63VYY252 pKa = 11.35GDD254 pKa = 5.47ADD256 pKa = 3.68NDD258 pKa = 4.39WIHH261 pKa = 6.97GEE263 pKa = 4.11GGHH266 pKa = 7.17DD267 pKa = 3.76EE268 pKa = 4.29LHH270 pKa = 6.72GGTGDD275 pKa = 3.46DD276 pKa = 4.15TIYY279 pKa = 11.24GGDD282 pKa = 4.04GYY284 pKa = 11.24DD285 pKa = 5.61LIWGDD290 pKa = 4.65DD291 pKa = 4.11GNDD294 pKa = 3.31TLYY297 pKa = 11.27GGNDD301 pKa = 3.76DD302 pKa = 3.7DD303 pKa = 4.98TIYY306 pKa = 11.07GGAGIDD312 pKa = 3.75VLYY315 pKa = 11.07GDD317 pKa = 5.3AGNDD321 pKa = 3.73TLSGDD326 pKa = 3.35AGGDD330 pKa = 3.36HH331 pKa = 7.45LYY333 pKa = 11.15GGTGNDD339 pKa = 3.42TMSGGTGNDD348 pKa = 2.65SFYY351 pKa = 11.46VLEE354 pKa = 4.81GDD356 pKa = 4.3GTSTMYY362 pKa = 11.24GDD364 pKa = 5.56DD365 pKa = 4.0DD366 pKa = 4.15WDD368 pKa = 4.08TVDD371 pKa = 4.65FAYY374 pKa = 9.53STGSAVDD381 pKa = 3.43VTYY384 pKa = 11.18SGDD387 pKa = 3.41NAFNFAFDD395 pKa = 3.96TGNSSGSGNSMEE407 pKa = 4.68NFILTGQNDD416 pKa = 4.05TVDD419 pKa = 3.63ASLDD423 pKa = 3.73ATGATYY429 pKa = 10.66DD430 pKa = 4.44LRR432 pKa = 11.84AGDD435 pKa = 3.89DD436 pKa = 3.71TFTGGSGNDD445 pKa = 3.14AVYY448 pKa = 10.61GGDD451 pKa = 3.63GADD454 pKa = 4.08TITLGAGGDD463 pKa = 3.93YY464 pKa = 11.39AEE466 pKa = 5.31GGAGNDD472 pKa = 4.05TIYY475 pKa = 11.44GDD477 pKa = 4.85AGEE480 pKa = 4.48DD481 pKa = 3.51WIEE484 pKa = 4.24GGTGNDD490 pKa = 3.32TLYY493 pKa = 11.28GGDD496 pKa = 3.73DD497 pKa = 3.94ADD499 pKa = 3.72TFFYY503 pKa = 10.99SDD505 pKa = 2.85GWGSDD510 pKa = 3.4TVFGGAGISDD520 pKa = 4.57ADD522 pKa = 3.78TLDD525 pKa = 3.7FSNVTTDD532 pKa = 3.25VTVTFSDD539 pKa = 3.89WEE541 pKa = 4.71DD542 pKa = 3.39GTATSGADD550 pKa = 2.98SVNFDD555 pKa = 3.95NIEE558 pKa = 4.4AIRR561 pKa = 11.84GGSGADD567 pKa = 3.73TIDD570 pKa = 3.79ASPDD574 pKa = 3.31GSGLLLDD581 pKa = 5.1GGDD584 pKa = 4.12GGDD587 pKa = 3.91TIIGGSGDD595 pKa = 3.73DD596 pKa = 3.85EE597 pKa = 5.88LYY599 pKa = 11.23GGDD602 pKa = 4.42GVDD605 pKa = 3.68TLFGGAGNDD614 pKa = 3.98AIDD617 pKa = 4.19GGRR620 pKa = 11.84GDD622 pKa = 3.86DD623 pKa = 4.44TITGGAGDD631 pKa = 4.12DD632 pKa = 4.62TITSSNGQDD641 pKa = 2.75TMVFADD647 pKa = 4.44GFGNDD652 pKa = 4.39IITDD656 pKa = 4.3FDD658 pKa = 4.07FTDD661 pKa = 4.05SNSDD665 pKa = 3.35GYY667 pKa = 9.63TDD669 pKa = 4.16DD670 pKa = 3.94QLDD673 pKa = 3.65VSALTDD679 pKa = 3.47SDD681 pKa = 4.3GNPVNAWDD689 pKa = 4.33VVVSDD694 pKa = 5.17DD695 pKa = 4.01GLGNAVLTFPNGEE708 pKa = 4.57SITLTGIAPTQVDD721 pKa = 4.55GAQQMHH727 pKa = 7.52AIGIPCFVSGTRR739 pKa = 11.84IMTPSGNRR747 pKa = 11.84LIEE750 pKa = 4.09EE751 pKa = 4.4LRR753 pKa = 11.84HH754 pKa = 5.69GDD756 pKa = 4.05LVSTLNSGAQPILWAGRR773 pKa = 11.84TTLKK777 pKa = 10.35RR778 pKa = 11.84ARR780 pKa = 11.84LALQPDD786 pKa = 4.75LLPIRR791 pKa = 11.84IRR793 pKa = 11.84DD794 pKa = 3.75GTLGNSGDD802 pKa = 3.7LWVSPQHH809 pKa = 6.59GLVLPDD815 pKa = 4.03LCSHH819 pKa = 6.81AKK821 pKa = 10.45AGVLVRR827 pKa = 11.84AKK829 pKa = 10.41HH830 pKa = 5.46LLEE833 pKa = 5.93HH834 pKa = 6.59GDD836 pKa = 3.36GRR838 pKa = 11.84VRR840 pKa = 11.84RR841 pKa = 11.84AKK843 pKa = 10.23GSRR846 pKa = 11.84DD847 pKa = 3.25VTYY850 pKa = 10.73HH851 pKa = 6.4HH852 pKa = 7.25LLLPRR857 pKa = 11.84HH858 pKa = 5.86ALILANGCASEE869 pKa = 4.26SLFPGRR875 pKa = 11.84FALSGFDD882 pKa = 3.19RR883 pKa = 11.84AAKK886 pKa = 10.62AEE888 pKa = 4.1LFDD891 pKa = 5.35LFPALEE897 pKa = 4.51TILRR901 pKa = 11.84SGAPATDD908 pKa = 3.61AARR911 pKa = 11.84MYY913 pKa = 8.49GQPVLRR919 pKa = 11.84FAKK922 pKa = 10.19GRR924 pKa = 11.84DD925 pKa = 2.85WRR927 pKa = 11.84AGHH930 pKa = 6.19IRR932 pKa = 11.84LPKK935 pKa = 9.96RR936 pKa = 11.84AA937 pKa = 3.85
Molecular weight: 96.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0ADR4|A0A0P0ADR4_9RHOB Sorbitol-6-phosphate 2-dehydrogenase OS=Celeribacter marinus OX=1397108 GN=IMCC12053_2686 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
909897 |
37 |
2082 |
290.3 |
31.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.03 ± 0.062 | 0.882 ± 0.014 |
6.405 ± 0.051 | 5.571 ± 0.042 |
3.811 ± 0.027 | 8.413 ± 0.046 |
2.114 ± 0.024 | 5.556 ± 0.033 |
3.68 ± 0.037 | 9.518 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.931 ± 0.024 | 2.837 ± 0.024 |
4.589 ± 0.034 | 3.194 ± 0.024 |
6.073 ± 0.041 | 5.471 ± 0.035 |
5.894 ± 0.037 | 7.476 ± 0.033 |
1.284 ± 0.018 | 2.272 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
