
Oceanotoga teriensis
Taxonomy: cellular organisms; Bacteria; Thermotogae; Thermotogae; Petrotogales; Petrotogaceae; Oceanotoga
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
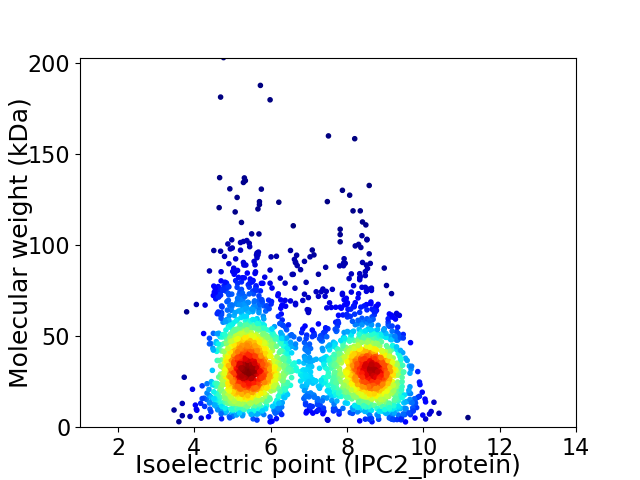
Virtual 2D-PAGE plot for 2638 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316D2T6|A0A316D2T6_9BACT Putative MATE family efflux protein OS=Oceanotoga teriensis OX=515440 GN=C7380_10770 PE=4 SV=1
MM1 pKa = 7.58SKK3 pKa = 10.72RR4 pKa = 11.84LTVLLFIIFTFILSFGSSYY23 pKa = 11.06TMNSFDD29 pKa = 3.81SSEE32 pKa = 4.09LNRR35 pKa = 11.84INSAMSTDD43 pKa = 3.41KK44 pKa = 11.57GNFSLNVAKK53 pKa = 10.62VIYY56 pKa = 10.21DD57 pKa = 3.63FYY59 pKa = 11.5NFDD62 pKa = 3.61WAGNEE67 pKa = 3.7YY68 pKa = 10.8LLRR71 pKa = 11.84DD72 pKa = 4.24LEE74 pKa = 5.72DD75 pKa = 3.16IMNLYY80 pKa = 10.62SEE82 pKa = 5.93DD83 pKa = 3.73IYY85 pKa = 11.66SEE87 pKa = 4.9DD88 pKa = 3.93FDD90 pKa = 6.24SEE92 pKa = 4.51FDD94 pKa = 4.46DD95 pKa = 4.29EE96 pKa = 6.17DD97 pKa = 4.02YY98 pKa = 11.52LEE100 pKa = 4.27KK101 pKa = 11.01LEE103 pKa = 5.12EE104 pKa = 4.14FFKK107 pKa = 10.97SLKK110 pKa = 10.8SSDD113 pKa = 4.12VYY115 pKa = 11.84DD116 pKa = 4.07LMVSIKK122 pKa = 10.9DD123 pKa = 3.55EE124 pKa = 4.86LKK126 pKa = 11.18DD127 pKa = 3.52LDD129 pKa = 4.26EE130 pKa = 5.22NKK132 pKa = 10.6DD133 pKa = 3.64SLVFLSKK140 pKa = 10.31EE141 pKa = 4.01AYY143 pKa = 9.94DD144 pKa = 4.76DD145 pKa = 3.78FVKK148 pKa = 10.89FINVDD153 pKa = 3.08SDD155 pKa = 3.97PYY157 pKa = 10.58FVEE160 pKa = 4.5FDD162 pKa = 3.84YY163 pKa = 10.1EE164 pKa = 4.63TVGNFAFDD172 pKa = 3.56YY173 pKa = 7.9RR174 pKa = 11.84TLKK177 pKa = 10.66ILDD180 pKa = 3.32ILANTFVYY188 pKa = 10.5MNDD191 pKa = 3.45NPSMLEE197 pKa = 3.83YY198 pKa = 10.15YY199 pKa = 10.76HH200 pKa = 6.74NFFSYY205 pKa = 10.76FEE207 pKa = 4.44SDD209 pKa = 3.36PDD211 pKa = 3.71EE212 pKa = 4.62EE213 pKa = 4.73DD214 pKa = 4.48LIRR217 pKa = 11.84PEE219 pKa = 4.2ANSVFSQMEE228 pKa = 4.07YY229 pKa = 10.9VEE231 pKa = 5.02PEE233 pKa = 4.04DD234 pKa = 5.65FEE236 pKa = 4.92PLLKK240 pKa = 10.6KK241 pKa = 10.48LAAVDD246 pKa = 5.07GDD248 pKa = 4.45SEE250 pKa = 4.64WISLRR255 pKa = 11.84DD256 pKa = 3.33IFIPEE261 pKa = 4.06ILILPPSYY269 pKa = 10.92EE270 pKa = 3.98YY271 pKa = 9.84FTISDD276 pKa = 3.59YY277 pKa = 11.96AMRR280 pKa = 11.84GMLLIDD286 pKa = 4.62DD287 pKa = 5.45FEE289 pKa = 4.77TSDD292 pKa = 3.58LFVNKK297 pKa = 9.78RR298 pKa = 11.84YY299 pKa = 10.37ADD301 pKa = 3.62LVDD304 pKa = 3.91YY305 pKa = 10.3FMNTSEE311 pKa = 5.21ADD313 pKa = 3.51DD314 pKa = 3.77FVYY317 pKa = 10.79YY318 pKa = 9.41YY319 pKa = 11.04YY320 pKa = 9.94PDD322 pKa = 3.29SYY324 pKa = 11.24GIKK327 pKa = 10.05EE328 pKa = 4.61DD329 pKa = 3.39FDD331 pKa = 4.25KK332 pKa = 11.34VLKK335 pKa = 10.32EE336 pKa = 4.13NYY338 pKa = 9.02NFRR341 pKa = 11.84YY342 pKa = 10.14ANPILGFLNVSFDD355 pKa = 3.77EE356 pKa = 4.73FLMEE360 pKa = 5.33EE361 pKa = 4.19DD362 pKa = 4.52LEE364 pKa = 5.43DD365 pKa = 3.53IVSEE369 pKa = 4.2FWMSINGVKK378 pKa = 10.26VYY380 pKa = 10.74NFRR383 pKa = 11.84DD384 pKa = 3.96FVEE387 pKa = 4.52EE388 pKa = 3.87FLNSYY393 pKa = 9.93KK394 pKa = 10.98DD395 pKa = 3.44VMNFKK400 pKa = 10.52FRR402 pKa = 11.84MMFNIDD408 pKa = 3.12MSTGYY413 pKa = 9.88EE414 pKa = 4.0EE415 pKa = 4.54PGEE418 pKa = 4.16VVEE421 pKa = 4.6PLVLGLDD428 pKa = 3.52MNMTFNYY435 pKa = 8.81PAYY438 pKa = 9.51KK439 pKa = 10.64DD440 pKa = 3.83PFDD443 pKa = 3.82VGAMNLRR450 pKa = 11.84FKK452 pKa = 11.11PEE454 pKa = 3.41IFNVVMSEE462 pKa = 4.04FAKK465 pKa = 10.87LEE467 pKa = 3.67QGYY470 pKa = 9.88FNISEE475 pKa = 4.13IVSNISEE482 pKa = 4.19YY483 pKa = 10.78TDD485 pKa = 3.61FLKK488 pKa = 11.09VEE490 pKa = 4.14NNDD493 pKa = 3.22LVFYY497 pKa = 10.73LNVDD501 pKa = 3.48NYY503 pKa = 10.17FDD505 pKa = 4.48SEE507 pKa = 4.11ISFVNFMSDD516 pKa = 4.36DD517 pKa = 3.58IVNLLSMFLMTAMSSDD533 pKa = 3.55YY534 pKa = 11.31NYY536 pKa = 11.59
MM1 pKa = 7.58SKK3 pKa = 10.72RR4 pKa = 11.84LTVLLFIIFTFILSFGSSYY23 pKa = 11.06TMNSFDD29 pKa = 3.81SSEE32 pKa = 4.09LNRR35 pKa = 11.84INSAMSTDD43 pKa = 3.41KK44 pKa = 11.57GNFSLNVAKK53 pKa = 10.62VIYY56 pKa = 10.21DD57 pKa = 3.63FYY59 pKa = 11.5NFDD62 pKa = 3.61WAGNEE67 pKa = 3.7YY68 pKa = 10.8LLRR71 pKa = 11.84DD72 pKa = 4.24LEE74 pKa = 5.72DD75 pKa = 3.16IMNLYY80 pKa = 10.62SEE82 pKa = 5.93DD83 pKa = 3.73IYY85 pKa = 11.66SEE87 pKa = 4.9DD88 pKa = 3.93FDD90 pKa = 6.24SEE92 pKa = 4.51FDD94 pKa = 4.46DD95 pKa = 4.29EE96 pKa = 6.17DD97 pKa = 4.02YY98 pKa = 11.52LEE100 pKa = 4.27KK101 pKa = 11.01LEE103 pKa = 5.12EE104 pKa = 4.14FFKK107 pKa = 10.97SLKK110 pKa = 10.8SSDD113 pKa = 4.12VYY115 pKa = 11.84DD116 pKa = 4.07LMVSIKK122 pKa = 10.9DD123 pKa = 3.55EE124 pKa = 4.86LKK126 pKa = 11.18DD127 pKa = 3.52LDD129 pKa = 4.26EE130 pKa = 5.22NKK132 pKa = 10.6DD133 pKa = 3.64SLVFLSKK140 pKa = 10.31EE141 pKa = 4.01AYY143 pKa = 9.94DD144 pKa = 4.76DD145 pKa = 3.78FVKK148 pKa = 10.89FINVDD153 pKa = 3.08SDD155 pKa = 3.97PYY157 pKa = 10.58FVEE160 pKa = 4.5FDD162 pKa = 3.84YY163 pKa = 10.1EE164 pKa = 4.63TVGNFAFDD172 pKa = 3.56YY173 pKa = 7.9RR174 pKa = 11.84TLKK177 pKa = 10.66ILDD180 pKa = 3.32ILANTFVYY188 pKa = 10.5MNDD191 pKa = 3.45NPSMLEE197 pKa = 3.83YY198 pKa = 10.15YY199 pKa = 10.76HH200 pKa = 6.74NFFSYY205 pKa = 10.76FEE207 pKa = 4.44SDD209 pKa = 3.36PDD211 pKa = 3.71EE212 pKa = 4.62EE213 pKa = 4.73DD214 pKa = 4.48LIRR217 pKa = 11.84PEE219 pKa = 4.2ANSVFSQMEE228 pKa = 4.07YY229 pKa = 10.9VEE231 pKa = 5.02PEE233 pKa = 4.04DD234 pKa = 5.65FEE236 pKa = 4.92PLLKK240 pKa = 10.6KK241 pKa = 10.48LAAVDD246 pKa = 5.07GDD248 pKa = 4.45SEE250 pKa = 4.64WISLRR255 pKa = 11.84DD256 pKa = 3.33IFIPEE261 pKa = 4.06ILILPPSYY269 pKa = 10.92EE270 pKa = 3.98YY271 pKa = 9.84FTISDD276 pKa = 3.59YY277 pKa = 11.96AMRR280 pKa = 11.84GMLLIDD286 pKa = 4.62DD287 pKa = 5.45FEE289 pKa = 4.77TSDD292 pKa = 3.58LFVNKK297 pKa = 9.78RR298 pKa = 11.84YY299 pKa = 10.37ADD301 pKa = 3.62LVDD304 pKa = 3.91YY305 pKa = 10.3FMNTSEE311 pKa = 5.21ADD313 pKa = 3.51DD314 pKa = 3.77FVYY317 pKa = 10.79YY318 pKa = 9.41YY319 pKa = 11.04YY320 pKa = 9.94PDD322 pKa = 3.29SYY324 pKa = 11.24GIKK327 pKa = 10.05EE328 pKa = 4.61DD329 pKa = 3.39FDD331 pKa = 4.25KK332 pKa = 11.34VLKK335 pKa = 10.32EE336 pKa = 4.13NYY338 pKa = 9.02NFRR341 pKa = 11.84YY342 pKa = 10.14ANPILGFLNVSFDD355 pKa = 3.77EE356 pKa = 4.73FLMEE360 pKa = 5.33EE361 pKa = 4.19DD362 pKa = 4.52LEE364 pKa = 5.43DD365 pKa = 3.53IVSEE369 pKa = 4.2FWMSINGVKK378 pKa = 10.26VYY380 pKa = 10.74NFRR383 pKa = 11.84DD384 pKa = 3.96FVEE387 pKa = 4.52EE388 pKa = 3.87FLNSYY393 pKa = 9.93KK394 pKa = 10.98DD395 pKa = 3.44VMNFKK400 pKa = 10.52FRR402 pKa = 11.84MMFNIDD408 pKa = 3.12MSTGYY413 pKa = 9.88EE414 pKa = 4.0EE415 pKa = 4.54PGEE418 pKa = 4.16VVEE421 pKa = 4.6PLVLGLDD428 pKa = 3.52MNMTFNYY435 pKa = 8.81PAYY438 pKa = 9.51KK439 pKa = 10.64DD440 pKa = 3.83PFDD443 pKa = 3.82VGAMNLRR450 pKa = 11.84FKK452 pKa = 11.11PEE454 pKa = 3.41IFNVVMSEE462 pKa = 4.04FAKK465 pKa = 10.87LEE467 pKa = 3.67QGYY470 pKa = 9.88FNISEE475 pKa = 4.13IVSNISEE482 pKa = 4.19YY483 pKa = 10.78TDD485 pKa = 3.61FLKK488 pKa = 11.09VEE490 pKa = 4.14NNDD493 pKa = 3.22LVFYY497 pKa = 10.73LNVDD501 pKa = 3.48NYY503 pKa = 10.17FDD505 pKa = 4.48SEE507 pKa = 4.11ISFVNFMSDD516 pKa = 4.36DD517 pKa = 3.58IVNLLSMFLMTAMSSDD533 pKa = 3.55YY534 pKa = 11.31NYY536 pKa = 11.59
Molecular weight: 63.44 kDa
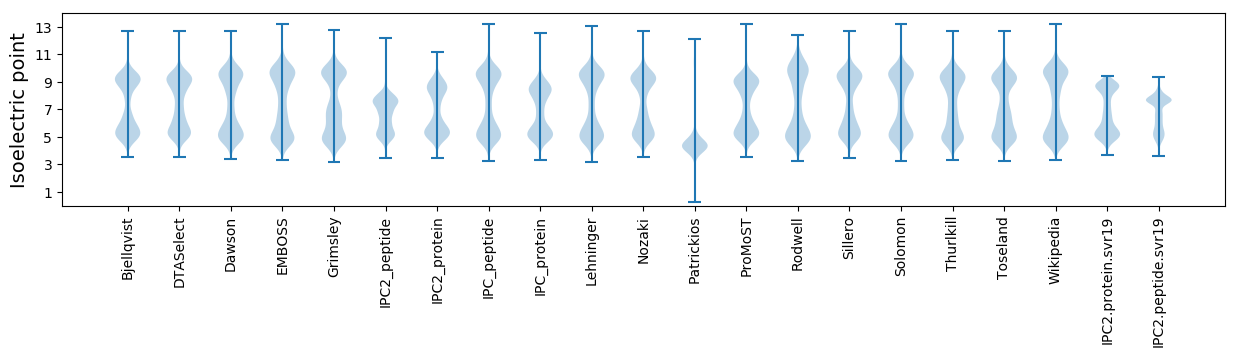
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316D0Q2|A0A316D0Q2_9BACT 3D (Asp-Asp-Asp) domain-containing protein OS=Oceanotoga teriensis OX=515440 GN=C7380_10290 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPSRR9 pKa = 11.84IKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 7.89RR14 pKa = 11.84THH16 pKa = 6.05GFLVRR21 pKa = 11.84KK22 pKa = 7.06RR23 pKa = 11.84TKK25 pKa = 10.3AGSSVLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPSRR9 pKa = 11.84IKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 7.89RR14 pKa = 11.84THH16 pKa = 6.05GFLVRR21 pKa = 11.84KK22 pKa = 7.06RR23 pKa = 11.84TKK25 pKa = 10.3AGSSVLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
858968 |
26 |
1762 |
325.6 |
37.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.394 ± 0.047 | 0.641 ± 0.014 |
5.608 ± 0.037 | 7.409 ± 0.051 |
5.524 ± 0.045 | 5.579 ± 0.048 |
1.243 ± 0.018 | 11.443 ± 0.062 |
10.147 ± 0.062 | 9.038 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.021 | 7.348 ± 0.056 |
2.805 ± 0.024 | 2.15 ± 0.025 |
2.768 ± 0.027 | 6.512 ± 0.038 |
4.439 ± 0.032 | 5.044 ± 0.042 |
0.754 ± 0.015 | 4.622 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
