
Crocosphaera watsonii WH 8501
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Aphanothecaceae; Crocosphaera; Crocosphaera watsonii
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
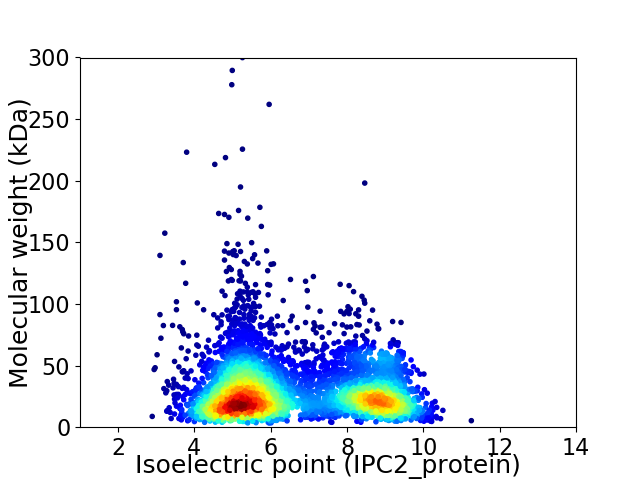
Virtual 2D-PAGE plot for 5659 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
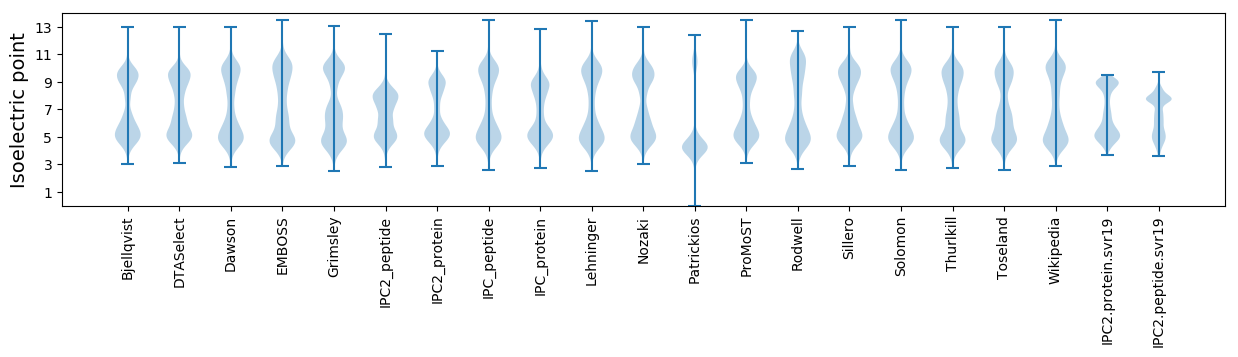
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4C6L7|Q4C6L7_CROWT Uncharacterized protein OS=Crocosphaera watsonii WH 8501 OX=165597 GN=CwatDRAFT_5003 PE=4 SV=1
MM1 pKa = 7.8LSTTFSGTTSYY12 pKa = 11.09SSSIAYY18 pKa = 9.6GFEE21 pKa = 3.82VEE23 pKa = 4.47NLINSSSSDD32 pKa = 3.4KK33 pKa = 10.81IYY35 pKa = 10.98LNNAANEE42 pKa = 4.02IMGYY46 pKa = 9.48NPNRR50 pKa = 11.84STGQDD55 pKa = 2.86KK56 pKa = 10.41IYY58 pKa = 10.85YY59 pKa = 7.55GTSQDD64 pKa = 3.96TLILDD69 pKa = 3.7YY70 pKa = 11.36AHH72 pKa = 6.82YY73 pKa = 10.28EE74 pKa = 4.13VTEE77 pKa = 4.24SQSGQNLIINLGGNGSVEE95 pKa = 4.08LVDD98 pKa = 5.8YY99 pKa = 11.12YY100 pKa = 11.63ADD102 pKa = 4.18PNNQIQISYY111 pKa = 9.64AIPPTPRR118 pKa = 11.84PEE120 pKa = 4.1PPEE123 pKa = 4.92GIDD126 pKa = 3.78IVIAEE131 pKa = 4.78IGQISNLDD139 pKa = 3.82HH140 pKa = 6.75NNQTIVLDD148 pKa = 3.74HH149 pKa = 7.21DD150 pKa = 4.09FVNPVILAQPLSFNDD165 pKa = 3.4GGVAIARR172 pKa = 11.84ITDD175 pKa = 3.38IQSDD179 pKa = 3.91RR180 pKa = 11.84FSVEE184 pKa = 3.22IQEE187 pKa = 4.7ASNEE191 pKa = 4.17DD192 pKa = 3.84GIHH195 pKa = 4.74TVEE198 pKa = 4.14TFSYY202 pKa = 10.47LVLEE206 pKa = 4.2EE207 pKa = 5.21GIWEE211 pKa = 4.35LSDD214 pKa = 3.2GTIIEE219 pKa = 4.16VGIINATNSSWSNINFSYY237 pKa = 11.15DD238 pKa = 3.34FVDD241 pKa = 4.5DD242 pKa = 4.4PVVLTQIQTNNDD254 pKa = 3.35SNFVKK259 pKa = 9.91TRR261 pKa = 11.84HH262 pKa = 5.8NNITQDD268 pKa = 3.18GFEE271 pKa = 4.29VLLEE275 pKa = 4.12KK276 pKa = 11.09DD277 pKa = 3.37EE278 pKa = 4.95ANNGTSYY285 pKa = 11.13GIEE288 pKa = 3.91TVAWMAISSGTGSWDD303 pKa = 3.05GNTFMAGNTGDD314 pKa = 3.63QVTHH318 pKa = 7.05DD319 pKa = 3.26WHH321 pKa = 6.81TIDD324 pKa = 5.68FGNAFNNTPKK334 pKa = 10.63FLGNIASYY342 pKa = 10.5YY343 pKa = 10.64GPDD346 pKa = 3.64PSGLRR351 pKa = 11.84YY352 pKa = 9.89QNLNNGNVQIKK363 pKa = 10.22IEE365 pKa = 4.37EE366 pKa = 4.24DD367 pKa = 3.11TSADD371 pKa = 3.56SEE373 pKa = 4.36IGHH376 pKa = 5.37TTEE379 pKa = 5.17DD380 pKa = 2.89VDD382 pKa = 3.95YY383 pKa = 10.93LAIEE387 pKa = 4.2ATGTLIGSTYY397 pKa = 11.16GLIPPTSNDD406 pKa = 3.85PIIAQLGQVTNLDD419 pKa = 4.27HH420 pKa = 6.98NNQTIVLDD428 pKa = 3.79HH429 pKa = 7.27DD430 pKa = 4.29FDD432 pKa = 4.83NPVIFANPLSYY443 pKa = 10.87NGPAPSIARR452 pKa = 11.84ITDD455 pKa = 3.14IQGDD459 pKa = 3.96RR460 pKa = 11.84FSVEE464 pKa = 3.65LQEE467 pKa = 4.91PSNEE471 pKa = 4.4DD472 pKa = 2.99GTHH475 pKa = 6.1AEE477 pKa = 4.13EE478 pKa = 4.43TVSFLALEE486 pKa = 4.06EE487 pKa = 5.19GIWEE491 pKa = 4.45LSDD494 pKa = 3.2GTIIEE499 pKa = 4.44VGTIDD504 pKa = 3.56TTHH507 pKa = 6.78SDD509 pKa = 2.49WHH511 pKa = 6.63NINFSYY517 pKa = 11.07DD518 pKa = 3.45FVDD521 pKa = 4.82GPVVLTQIQTNNDD534 pKa = 2.75SSFVKK539 pKa = 9.58TRR541 pKa = 11.84HH542 pKa = 5.81NNITQDD548 pKa = 3.18GFEE551 pKa = 4.28VLLEE555 pKa = 4.07NDD557 pKa = 3.89EE558 pKa = 5.32ANMNSGHH565 pKa = 6.01GNEE568 pKa = 4.16TVAWMAISSGTGSWDD583 pKa = 3.05GNTFMAGNTGDD594 pKa = 3.63QVTHH598 pKa = 7.05DD599 pKa = 3.26WHH601 pKa = 6.81TIDD604 pKa = 5.68FGNAFNNTPKK614 pKa = 10.63FLGNIASYY622 pKa = 10.5YY623 pKa = 10.64GPDD626 pKa = 3.64PSGLRR631 pKa = 11.84YY632 pKa = 9.89QNLNNGNVEE641 pKa = 4.0IKK643 pKa = 10.13IEE645 pKa = 4.0EE646 pKa = 4.97DD647 pKa = 2.83ISIDD651 pKa = 3.82EE652 pKa = 4.53EE653 pKa = 4.51VTHH656 pKa = 5.95ITEE659 pKa = 4.67DD660 pKa = 3.54VHH662 pKa = 7.86FLAIEE667 pKa = 4.26GTGTLTGSTYY677 pKa = 10.2IDD679 pKa = 4.63PDD681 pKa = 3.63NDD683 pKa = 4.03PDD685 pKa = 4.1PVSTIAQVGQITNLDD700 pKa = 3.98EE701 pKa = 4.48NNQTIVLDD709 pKa = 3.79HH710 pKa = 7.27DD711 pKa = 4.29FDD713 pKa = 4.83NPVIFANPLSYY724 pKa = 10.87NGPAPSIARR733 pKa = 11.84ITDD736 pKa = 3.21IQSDD740 pKa = 3.94RR741 pKa = 11.84FSVEE745 pKa = 3.44LQEE748 pKa = 4.91PSNEE752 pKa = 4.4DD753 pKa = 2.99GTHH756 pKa = 6.02AEE758 pKa = 4.15EE759 pKa = 4.46TFSFLALEE767 pKa = 4.36KK768 pKa = 10.77GVWTLSDD775 pKa = 3.32GTVIEE780 pKa = 4.65VGTIDD785 pKa = 3.37TNAIAGSYY793 pKa = 8.56WEE795 pKa = 5.48NITFDD800 pKa = 3.98YY801 pKa = 11.15DD802 pKa = 3.36FTNAPIVLTQVQTDD816 pKa = 3.25NDD818 pKa = 3.47ASFVKK823 pKa = 9.68TRR825 pKa = 11.84QNNITQDD832 pKa = 3.56GFDD835 pKa = 3.85LALEE839 pKa = 4.11NDD841 pKa = 3.82EE842 pKa = 5.55ANLNSGHH849 pKa = 5.25GTEE852 pKa = 4.35TVARR856 pKa = 11.84VAISSGTGDD865 pKa = 3.09WDD867 pKa = 3.85GNTFMAGEE875 pKa = 4.16TGDD878 pKa = 3.84YY879 pKa = 8.78VTEE882 pKa = 4.19AFYY885 pKa = 10.46TLNFGNAFNKK895 pKa = 10.1APKK898 pKa = 9.98FLGNIASYY906 pKa = 10.61YY907 pKa = 10.66GSDD910 pKa = 3.63PSGLRR915 pKa = 11.84YY916 pKa = 9.88QNLNNGNVEE925 pKa = 4.04IKK927 pKa = 10.23IEE929 pKa = 4.17EE930 pKa = 4.15DD931 pKa = 3.18TSIDD935 pKa = 3.48EE936 pKa = 4.65EE937 pKa = 4.71IIHH940 pKa = 5.73ITEE943 pKa = 4.11NVHH946 pKa = 6.4FLAIEE951 pKa = 4.24GTGTLTGSANTGNNDD966 pKa = 3.95PLTGLATEE974 pKa = 4.33QTATASQDD982 pKa = 2.79IFVVGNAQEE991 pKa = 4.27PLYY994 pKa = 9.95DD995 pKa = 3.3TYY997 pKa = 11.74GKK999 pKa = 9.87HH1000 pKa = 7.32DD1001 pKa = 4.01YY1002 pKa = 11.46LEE1004 pKa = 4.25ILGFDD1009 pKa = 3.28QSEE1012 pKa = 4.56DD1013 pKa = 3.72VIQLNGIADD1022 pKa = 4.11NYY1024 pKa = 11.03SLGASPFDD1032 pKa = 4.96SNDD1035 pKa = 2.84QGIFLKK1041 pKa = 10.67VAGMQDD1047 pKa = 3.35EE1048 pKa = 4.53LVAIVKK1054 pKa = 10.24DD1055 pKa = 4.09NNNLDD1060 pKa = 3.93LNSNQFVFVV1069 pKa = 3.68
MM1 pKa = 7.8LSTTFSGTTSYY12 pKa = 11.09SSSIAYY18 pKa = 9.6GFEE21 pKa = 3.82VEE23 pKa = 4.47NLINSSSSDD32 pKa = 3.4KK33 pKa = 10.81IYY35 pKa = 10.98LNNAANEE42 pKa = 4.02IMGYY46 pKa = 9.48NPNRR50 pKa = 11.84STGQDD55 pKa = 2.86KK56 pKa = 10.41IYY58 pKa = 10.85YY59 pKa = 7.55GTSQDD64 pKa = 3.96TLILDD69 pKa = 3.7YY70 pKa = 11.36AHH72 pKa = 6.82YY73 pKa = 10.28EE74 pKa = 4.13VTEE77 pKa = 4.24SQSGQNLIINLGGNGSVEE95 pKa = 4.08LVDD98 pKa = 5.8YY99 pKa = 11.12YY100 pKa = 11.63ADD102 pKa = 4.18PNNQIQISYY111 pKa = 9.64AIPPTPRR118 pKa = 11.84PEE120 pKa = 4.1PPEE123 pKa = 4.92GIDD126 pKa = 3.78IVIAEE131 pKa = 4.78IGQISNLDD139 pKa = 3.82HH140 pKa = 6.75NNQTIVLDD148 pKa = 3.74HH149 pKa = 7.21DD150 pKa = 4.09FVNPVILAQPLSFNDD165 pKa = 3.4GGVAIARR172 pKa = 11.84ITDD175 pKa = 3.38IQSDD179 pKa = 3.91RR180 pKa = 11.84FSVEE184 pKa = 3.22IQEE187 pKa = 4.7ASNEE191 pKa = 4.17DD192 pKa = 3.84GIHH195 pKa = 4.74TVEE198 pKa = 4.14TFSYY202 pKa = 10.47LVLEE206 pKa = 4.2EE207 pKa = 5.21GIWEE211 pKa = 4.35LSDD214 pKa = 3.2GTIIEE219 pKa = 4.16VGIINATNSSWSNINFSYY237 pKa = 11.15DD238 pKa = 3.34FVDD241 pKa = 4.5DD242 pKa = 4.4PVVLTQIQTNNDD254 pKa = 3.35SNFVKK259 pKa = 9.91TRR261 pKa = 11.84HH262 pKa = 5.8NNITQDD268 pKa = 3.18GFEE271 pKa = 4.29VLLEE275 pKa = 4.12KK276 pKa = 11.09DD277 pKa = 3.37EE278 pKa = 4.95ANNGTSYY285 pKa = 11.13GIEE288 pKa = 3.91TVAWMAISSGTGSWDD303 pKa = 3.05GNTFMAGNTGDD314 pKa = 3.63QVTHH318 pKa = 7.05DD319 pKa = 3.26WHH321 pKa = 6.81TIDD324 pKa = 5.68FGNAFNNTPKK334 pKa = 10.63FLGNIASYY342 pKa = 10.5YY343 pKa = 10.64GPDD346 pKa = 3.64PSGLRR351 pKa = 11.84YY352 pKa = 9.89QNLNNGNVQIKK363 pKa = 10.22IEE365 pKa = 4.37EE366 pKa = 4.24DD367 pKa = 3.11TSADD371 pKa = 3.56SEE373 pKa = 4.36IGHH376 pKa = 5.37TTEE379 pKa = 5.17DD380 pKa = 2.89VDD382 pKa = 3.95YY383 pKa = 10.93LAIEE387 pKa = 4.2ATGTLIGSTYY397 pKa = 11.16GLIPPTSNDD406 pKa = 3.85PIIAQLGQVTNLDD419 pKa = 4.27HH420 pKa = 6.98NNQTIVLDD428 pKa = 3.79HH429 pKa = 7.27DD430 pKa = 4.29FDD432 pKa = 4.83NPVIFANPLSYY443 pKa = 10.87NGPAPSIARR452 pKa = 11.84ITDD455 pKa = 3.14IQGDD459 pKa = 3.96RR460 pKa = 11.84FSVEE464 pKa = 3.65LQEE467 pKa = 4.91PSNEE471 pKa = 4.4DD472 pKa = 2.99GTHH475 pKa = 6.1AEE477 pKa = 4.13EE478 pKa = 4.43TVSFLALEE486 pKa = 4.06EE487 pKa = 5.19GIWEE491 pKa = 4.45LSDD494 pKa = 3.2GTIIEE499 pKa = 4.44VGTIDD504 pKa = 3.56TTHH507 pKa = 6.78SDD509 pKa = 2.49WHH511 pKa = 6.63NINFSYY517 pKa = 11.07DD518 pKa = 3.45FVDD521 pKa = 4.82GPVVLTQIQTNNDD534 pKa = 2.75SSFVKK539 pKa = 9.58TRR541 pKa = 11.84HH542 pKa = 5.81NNITQDD548 pKa = 3.18GFEE551 pKa = 4.28VLLEE555 pKa = 4.07NDD557 pKa = 3.89EE558 pKa = 5.32ANMNSGHH565 pKa = 6.01GNEE568 pKa = 4.16TVAWMAISSGTGSWDD583 pKa = 3.05GNTFMAGNTGDD594 pKa = 3.63QVTHH598 pKa = 7.05DD599 pKa = 3.26WHH601 pKa = 6.81TIDD604 pKa = 5.68FGNAFNNTPKK614 pKa = 10.63FLGNIASYY622 pKa = 10.5YY623 pKa = 10.64GPDD626 pKa = 3.64PSGLRR631 pKa = 11.84YY632 pKa = 9.89QNLNNGNVEE641 pKa = 4.0IKK643 pKa = 10.13IEE645 pKa = 4.0EE646 pKa = 4.97DD647 pKa = 2.83ISIDD651 pKa = 3.82EE652 pKa = 4.53EE653 pKa = 4.51VTHH656 pKa = 5.95ITEE659 pKa = 4.67DD660 pKa = 3.54VHH662 pKa = 7.86FLAIEE667 pKa = 4.26GTGTLTGSTYY677 pKa = 10.2IDD679 pKa = 4.63PDD681 pKa = 3.63NDD683 pKa = 4.03PDD685 pKa = 4.1PVSTIAQVGQITNLDD700 pKa = 3.98EE701 pKa = 4.48NNQTIVLDD709 pKa = 3.79HH710 pKa = 7.27DD711 pKa = 4.29FDD713 pKa = 4.83NPVIFANPLSYY724 pKa = 10.87NGPAPSIARR733 pKa = 11.84ITDD736 pKa = 3.21IQSDD740 pKa = 3.94RR741 pKa = 11.84FSVEE745 pKa = 3.44LQEE748 pKa = 4.91PSNEE752 pKa = 4.4DD753 pKa = 2.99GTHH756 pKa = 6.02AEE758 pKa = 4.15EE759 pKa = 4.46TFSFLALEE767 pKa = 4.36KK768 pKa = 10.77GVWTLSDD775 pKa = 3.32GTVIEE780 pKa = 4.65VGTIDD785 pKa = 3.37TNAIAGSYY793 pKa = 8.56WEE795 pKa = 5.48NITFDD800 pKa = 3.98YY801 pKa = 11.15DD802 pKa = 3.36FTNAPIVLTQVQTDD816 pKa = 3.25NDD818 pKa = 3.47ASFVKK823 pKa = 9.68TRR825 pKa = 11.84QNNITQDD832 pKa = 3.56GFDD835 pKa = 3.85LALEE839 pKa = 4.11NDD841 pKa = 3.82EE842 pKa = 5.55ANLNSGHH849 pKa = 5.25GTEE852 pKa = 4.35TVARR856 pKa = 11.84VAISSGTGDD865 pKa = 3.09WDD867 pKa = 3.85GNTFMAGEE875 pKa = 4.16TGDD878 pKa = 3.84YY879 pKa = 8.78VTEE882 pKa = 4.19AFYY885 pKa = 10.46TLNFGNAFNKK895 pKa = 10.1APKK898 pKa = 9.98FLGNIASYY906 pKa = 10.61YY907 pKa = 10.66GSDD910 pKa = 3.63PSGLRR915 pKa = 11.84YY916 pKa = 9.88QNLNNGNVEE925 pKa = 4.04IKK927 pKa = 10.23IEE929 pKa = 4.17EE930 pKa = 4.15DD931 pKa = 3.18TSIDD935 pKa = 3.48EE936 pKa = 4.65EE937 pKa = 4.71IIHH940 pKa = 5.73ITEE943 pKa = 4.11NVHH946 pKa = 6.4FLAIEE951 pKa = 4.24GTGTLTGSANTGNNDD966 pKa = 3.95PLTGLATEE974 pKa = 4.33QTATASQDD982 pKa = 2.79IFVVGNAQEE991 pKa = 4.27PLYY994 pKa = 9.95DD995 pKa = 3.3TYY997 pKa = 11.74GKK999 pKa = 9.87HH1000 pKa = 7.32DD1001 pKa = 4.01YY1002 pKa = 11.46LEE1004 pKa = 4.25ILGFDD1009 pKa = 3.28QSEE1012 pKa = 4.56DD1013 pKa = 3.72VIQLNGIADD1022 pKa = 4.11NYY1024 pKa = 11.03SLGASPFDD1032 pKa = 4.96SNDD1035 pKa = 2.84QGIFLKK1041 pKa = 10.67VAGMQDD1047 pKa = 3.35EE1048 pKa = 4.53LVAIVKK1054 pKa = 10.24DD1055 pKa = 4.09NNNLDD1060 pKa = 3.93LNSNQFVFVV1069 pKa = 3.68
Molecular weight: 116.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4C5Z7|Q4C5Z7_CROWT Uncharacterized protein OS=Crocosphaera watsonii WH 8501 OX=165597 GN=CwatDRAFT_4741 PE=4 SV=1
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84KK25 pKa = 9.23SNGRR29 pKa = 11.84KK30 pKa = 9.11VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.98KK38 pKa = 9.65GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84KK25 pKa = 9.23SNGRR29 pKa = 11.84KK30 pKa = 9.11VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.98KK38 pKa = 9.65GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1581675 |
29 |
2652 |
279.5 |
31.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.094 ± 0.037 | 1.141 ± 0.012 |
5.045 ± 0.032 | 6.95 ± 0.033 |
4.148 ± 0.025 | 6.178 ± 0.038 |
1.84 ± 0.015 | 7.471 ± 0.033 |
6.791 ± 0.05 | 10.725 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.848 ± 0.016 | 4.926 ± 0.03 |
4.403 ± 0.026 | 4.879 ± 0.025 |
4.916 ± 0.029 | 6.483 ± 0.024 |
5.421 ± 0.027 | 5.947 ± 0.027 |
1.429 ± 0.015 | 3.363 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
