
Streptohalobacillus salinus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Streptohalobacillus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
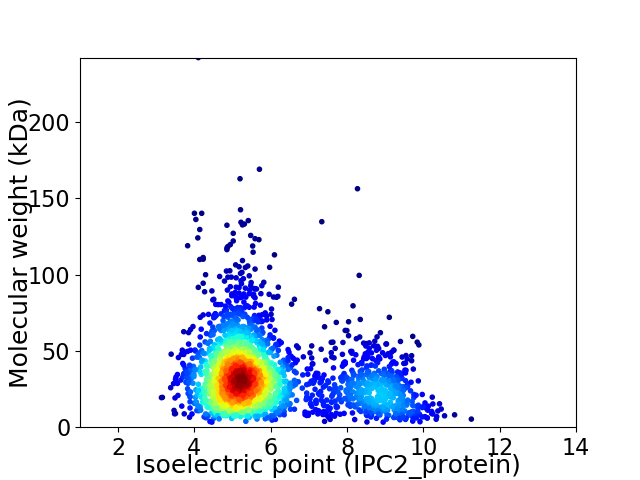
Virtual 2D-PAGE plot for 2307 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V3W2J6|A0A2V3W2J6_9BACI Putative MATE family efflux protein OS=Streptohalobacillus salinus OX=621096 GN=DES38_11625 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 10.24KK3 pKa = 10.25VLGVLVTLLLLITLAACGGSEE24 pKa = 5.38DD25 pKa = 4.97EE26 pKa = 5.19DD27 pKa = 4.0TSGDD31 pKa = 3.49NGGEE35 pKa = 4.29GTEE38 pKa = 4.13SDD40 pKa = 4.02GPTEE44 pKa = 4.03VTAWAWDD51 pKa = 3.58PNFNIRR57 pKa = 11.84ALEE60 pKa = 4.06LAEE63 pKa = 3.91EE64 pKa = 4.81AYY66 pKa = 10.58DD67 pKa = 5.15GEE69 pKa = 4.69TEE71 pKa = 4.23LDD73 pKa = 3.69LEE75 pKa = 4.76IIEE78 pKa = 4.59NAQDD82 pKa = 5.44DD83 pKa = 4.02IVQRR87 pKa = 11.84LNTGLSSGTTQGMPNIVLIEE107 pKa = 4.37DD108 pKa = 3.41YY109 pKa = 10.36RR110 pKa = 11.84AQSFLQSYY118 pKa = 9.92PEE120 pKa = 3.98AFYY123 pKa = 10.7PLTDD127 pKa = 4.42YY128 pKa = 11.31INPDD132 pKa = 3.23DD133 pKa = 3.9FASYY137 pKa = 10.27KK138 pKa = 10.49VEE140 pKa = 3.71ATSFDD145 pKa = 3.62GEE147 pKa = 4.45NYY149 pKa = 10.51GLPFDD154 pKa = 4.35SGVTGLYY161 pKa = 10.41VRR163 pKa = 11.84TDD165 pKa = 3.59YY166 pKa = 11.53LEE168 pKa = 4.16EE169 pKa = 5.52AGYY172 pKa = 8.16TVEE175 pKa = 5.62DD176 pKa = 3.72VTDD179 pKa = 3.9ITWSEE184 pKa = 4.13YY185 pKa = 10.38IEE187 pKa = 4.13IGKK190 pKa = 9.29DD191 pKa = 3.17VKK193 pKa = 10.77EE194 pKa = 3.86ATGYY198 pKa = 11.69NMITMDD204 pKa = 4.55PNDD207 pKa = 3.83FGLVRR212 pKa = 11.84MMLQSAGSWYY222 pKa = 10.51FEE224 pKa = 4.46EE225 pKa = 6.13DD226 pKa = 3.17GSTPNLEE233 pKa = 4.17GNPALRR239 pKa = 11.84EE240 pKa = 3.83AFEE243 pKa = 3.97VHH245 pKa = 6.0KK246 pKa = 11.24AMMEE250 pKa = 3.73ADD252 pKa = 3.33IVRR255 pKa = 11.84LHH257 pKa = 6.76SDD259 pKa = 2.93WSQFVGGFNSGDD271 pKa = 3.62VASVPTGNWITPSVKK286 pKa = 10.46AEE288 pKa = 3.74EE289 pKa = 4.31SQAGNWAVVPQPRR302 pKa = 11.84LEE304 pKa = 4.13VDD306 pKa = 3.44GGTTASNLGGSSWYY320 pKa = 10.37VLNTDD325 pKa = 3.43GKK327 pKa = 8.83EE328 pKa = 3.7AAAEE332 pKa = 3.96FLANTFGSNEE342 pKa = 3.83EE343 pKa = 4.47FYY345 pKa = 8.45TTLVEE350 pKa = 4.56EE351 pKa = 4.57IGALGTYY358 pKa = 9.31TPGTQGEE365 pKa = 4.21AFEE368 pKa = 5.78AGDD371 pKa = 4.11EE372 pKa = 4.35FFDD375 pKa = 4.27GQSIIQDD382 pKa = 3.47FSNWTEE388 pKa = 4.02EE389 pKa = 4.15IPPVNYY395 pKa = 10.31GMHH398 pKa = 6.84TYY400 pKa = 10.54AIDD403 pKa = 5.26DD404 pKa = 3.7ILVVEE409 pKa = 4.4MQHH412 pKa = 6.39YY413 pKa = 10.83LDD415 pKa = 4.92GRR417 pKa = 11.84SLDD420 pKa = 3.81EE421 pKa = 4.69ALSNAQDD428 pKa = 3.43QAEE431 pKa = 4.35TQLRR435 pKa = 3.83
MM1 pKa = 7.34KK2 pKa = 10.24KK3 pKa = 10.25VLGVLVTLLLLITLAACGGSEE24 pKa = 5.38DD25 pKa = 4.97EE26 pKa = 5.19DD27 pKa = 4.0TSGDD31 pKa = 3.49NGGEE35 pKa = 4.29GTEE38 pKa = 4.13SDD40 pKa = 4.02GPTEE44 pKa = 4.03VTAWAWDD51 pKa = 3.58PNFNIRR57 pKa = 11.84ALEE60 pKa = 4.06LAEE63 pKa = 3.91EE64 pKa = 4.81AYY66 pKa = 10.58DD67 pKa = 5.15GEE69 pKa = 4.69TEE71 pKa = 4.23LDD73 pKa = 3.69LEE75 pKa = 4.76IIEE78 pKa = 4.59NAQDD82 pKa = 5.44DD83 pKa = 4.02IVQRR87 pKa = 11.84LNTGLSSGTTQGMPNIVLIEE107 pKa = 4.37DD108 pKa = 3.41YY109 pKa = 10.36RR110 pKa = 11.84AQSFLQSYY118 pKa = 9.92PEE120 pKa = 3.98AFYY123 pKa = 10.7PLTDD127 pKa = 4.42YY128 pKa = 11.31INPDD132 pKa = 3.23DD133 pKa = 3.9FASYY137 pKa = 10.27KK138 pKa = 10.49VEE140 pKa = 3.71ATSFDD145 pKa = 3.62GEE147 pKa = 4.45NYY149 pKa = 10.51GLPFDD154 pKa = 4.35SGVTGLYY161 pKa = 10.41VRR163 pKa = 11.84TDD165 pKa = 3.59YY166 pKa = 11.53LEE168 pKa = 4.16EE169 pKa = 5.52AGYY172 pKa = 8.16TVEE175 pKa = 5.62DD176 pKa = 3.72VTDD179 pKa = 3.9ITWSEE184 pKa = 4.13YY185 pKa = 10.38IEE187 pKa = 4.13IGKK190 pKa = 9.29DD191 pKa = 3.17VKK193 pKa = 10.77EE194 pKa = 3.86ATGYY198 pKa = 11.69NMITMDD204 pKa = 4.55PNDD207 pKa = 3.83FGLVRR212 pKa = 11.84MMLQSAGSWYY222 pKa = 10.51FEE224 pKa = 4.46EE225 pKa = 6.13DD226 pKa = 3.17GSTPNLEE233 pKa = 4.17GNPALRR239 pKa = 11.84EE240 pKa = 3.83AFEE243 pKa = 3.97VHH245 pKa = 6.0KK246 pKa = 11.24AMMEE250 pKa = 3.73ADD252 pKa = 3.33IVRR255 pKa = 11.84LHH257 pKa = 6.76SDD259 pKa = 2.93WSQFVGGFNSGDD271 pKa = 3.62VASVPTGNWITPSVKK286 pKa = 10.46AEE288 pKa = 3.74EE289 pKa = 4.31SQAGNWAVVPQPRR302 pKa = 11.84LEE304 pKa = 4.13VDD306 pKa = 3.44GGTTASNLGGSSWYY320 pKa = 10.37VLNTDD325 pKa = 3.43GKK327 pKa = 8.83EE328 pKa = 3.7AAAEE332 pKa = 3.96FLANTFGSNEE342 pKa = 3.83EE343 pKa = 4.47FYY345 pKa = 8.45TTLVEE350 pKa = 4.56EE351 pKa = 4.57IGALGTYY358 pKa = 9.31TPGTQGEE365 pKa = 4.21AFEE368 pKa = 5.78AGDD371 pKa = 4.11EE372 pKa = 4.35FFDD375 pKa = 4.27GQSIIQDD382 pKa = 3.47FSNWTEE388 pKa = 4.02EE389 pKa = 4.15IPPVNYY395 pKa = 10.31GMHH398 pKa = 6.84TYY400 pKa = 10.54AIDD403 pKa = 5.26DD404 pKa = 3.7ILVVEE409 pKa = 4.4MQHH412 pKa = 6.39YY413 pKa = 10.83LDD415 pKa = 4.92GRR417 pKa = 11.84SLDD420 pKa = 3.81EE421 pKa = 4.69ALSNAQDD428 pKa = 3.43QAEE431 pKa = 4.35TQLRR435 pKa = 3.83
Molecular weight: 47.7 kDa
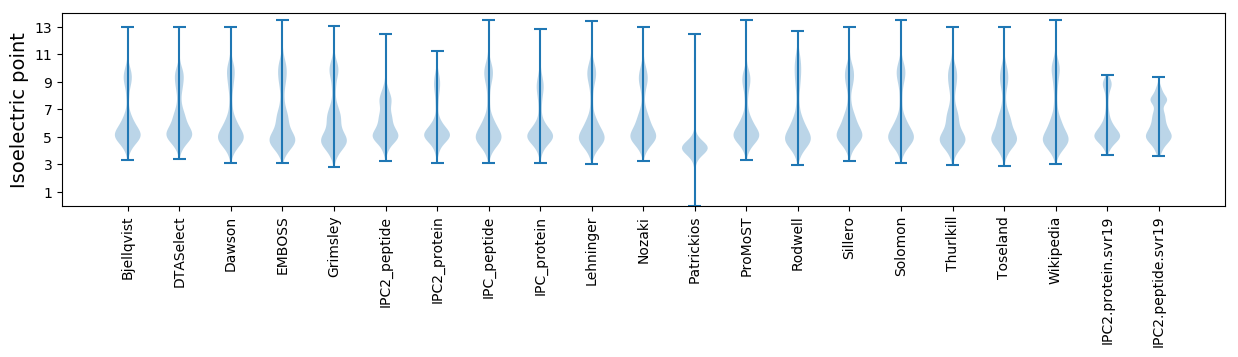
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V3WQH4|A0A2V3WQH4_9BACI Transketolase OS=Streptohalobacillus salinus OX=621096 GN=DES38_10781 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.02KK14 pKa = 8.47VHH16 pKa = 5.57GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 10.2NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.51KK37 pKa = 10.32GRR39 pKa = 11.84RR40 pKa = 11.84VLSAA44 pKa = 3.76
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.02KK14 pKa = 8.47VHH16 pKa = 5.57GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 10.2NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.51KK37 pKa = 10.32GRR39 pKa = 11.84RR40 pKa = 11.84VLSAA44 pKa = 3.76
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
724221 |
29 |
2174 |
313.9 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.377 ± 0.045 | 0.525 ± 0.012 |
5.983 ± 0.044 | 7.387 ± 0.052 |
4.421 ± 0.039 | 6.41 ± 0.042 |
2.323 ± 0.025 | 7.577 ± 0.049 |
6.144 ± 0.054 | 9.844 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.719 ± 0.019 | 4.166 ± 0.038 |
3.405 ± 0.027 | 4.179 ± 0.037 |
4.191 ± 0.031 | 5.571 ± 0.037 |
6.04 ± 0.033 | 7.275 ± 0.036 |
0.833 ± 0.018 | 3.631 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
