
Changjiang tombus-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
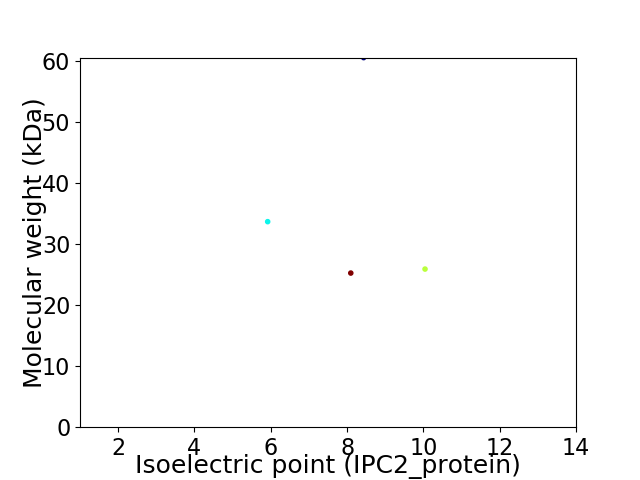
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
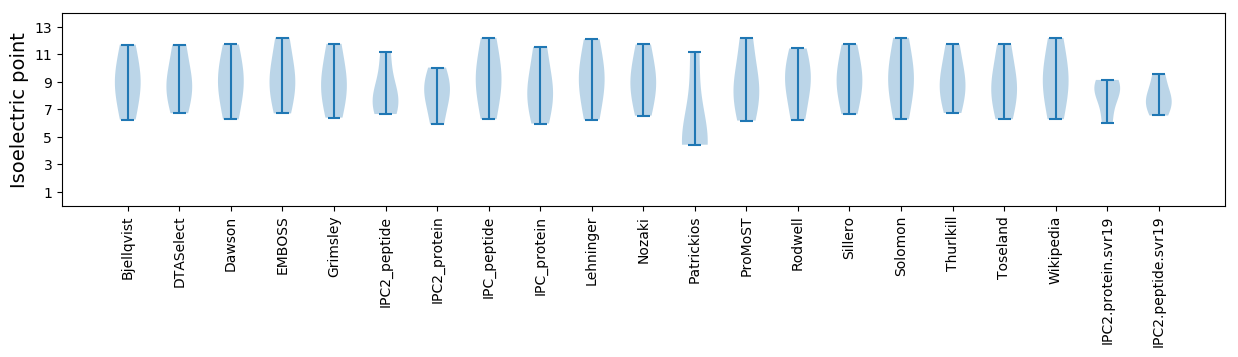
Protein with the lowest isoelectric point:
>tr|A0A1L3KFW5|A0A1L3KFW5_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 3 OX=1922817 PE=4 SV=1
MM1 pKa = 7.81ALRR4 pKa = 11.84HH5 pKa = 5.31LRR7 pKa = 11.84RR8 pKa = 11.84WLQLDD13 pKa = 3.62YY14 pKa = 11.06TPGGGVRR21 pKa = 11.84SRR23 pKa = 11.84RR24 pKa = 11.84EE25 pKa = 3.88CEE27 pKa = 3.35AHH29 pKa = 6.84YY30 pKa = 10.94GEE32 pKa = 4.87EE33 pKa = 3.95WSILIAQEE41 pKa = 3.74RR42 pKa = 11.84MTRR45 pKa = 11.84EE46 pKa = 3.7HH47 pKa = 7.21ASNAKK52 pKa = 9.96DD53 pKa = 3.19WFLGTAPCDD62 pKa = 3.48LLLPQIVVAPQPSEE76 pKa = 4.01GNAAAPAALSRR87 pKa = 11.84SEE89 pKa = 4.57DD90 pKa = 3.47SVDD93 pKa = 3.49AVSEE97 pKa = 4.37SSDD100 pKa = 3.58SVPDD104 pKa = 3.58VPGAPTPATALLVGSIPVTCPLVTDD129 pKa = 4.51RR130 pKa = 11.84PSAVLSPPTSHH141 pKa = 6.91TGAPTPPAASASRR154 pKa = 11.84VNTLCRR160 pKa = 11.84LALGVHH166 pKa = 6.66PGDD169 pKa = 4.17LTWVPAVTSTAIQVSRR185 pKa = 11.84TLVEE189 pKa = 4.01GTRR192 pKa = 11.84RR193 pKa = 11.84ATAQARR199 pKa = 11.84GWIRR203 pKa = 11.84AAVAVAEE210 pKa = 4.83RR211 pKa = 11.84EE212 pKa = 4.34LCPPPALEE220 pKa = 4.01EE221 pKa = 4.15LKK223 pKa = 10.62PVAKK227 pKa = 10.38GPRR230 pKa = 11.84QEE232 pKa = 4.07VGEE235 pKa = 4.05AASWVNAEE243 pKa = 4.19SVAMEE248 pKa = 4.07LRR250 pKa = 11.84ALFGLLTDD258 pKa = 4.19TPVHH262 pKa = 6.51RR263 pKa = 11.84EE264 pKa = 3.42MGGRR268 pKa = 11.84VARR271 pKa = 11.84EE272 pKa = 3.67ILRR275 pKa = 11.84DD276 pKa = 3.22RR277 pKa = 11.84CRR279 pKa = 11.84CGRR282 pKa = 11.84EE283 pKa = 3.98DD284 pKa = 3.03TWYY287 pKa = 10.81LSTSAVTHH295 pKa = 6.2WLSPTLTDD303 pKa = 4.14LVLATRR309 pKa = 11.84PQGFHH314 pKa = 6.35
MM1 pKa = 7.81ALRR4 pKa = 11.84HH5 pKa = 5.31LRR7 pKa = 11.84RR8 pKa = 11.84WLQLDD13 pKa = 3.62YY14 pKa = 11.06TPGGGVRR21 pKa = 11.84SRR23 pKa = 11.84RR24 pKa = 11.84EE25 pKa = 3.88CEE27 pKa = 3.35AHH29 pKa = 6.84YY30 pKa = 10.94GEE32 pKa = 4.87EE33 pKa = 3.95WSILIAQEE41 pKa = 3.74RR42 pKa = 11.84MTRR45 pKa = 11.84EE46 pKa = 3.7HH47 pKa = 7.21ASNAKK52 pKa = 9.96DD53 pKa = 3.19WFLGTAPCDD62 pKa = 3.48LLLPQIVVAPQPSEE76 pKa = 4.01GNAAAPAALSRR87 pKa = 11.84SEE89 pKa = 4.57DD90 pKa = 3.47SVDD93 pKa = 3.49AVSEE97 pKa = 4.37SSDD100 pKa = 3.58SVPDD104 pKa = 3.58VPGAPTPATALLVGSIPVTCPLVTDD129 pKa = 4.51RR130 pKa = 11.84PSAVLSPPTSHH141 pKa = 6.91TGAPTPPAASASRR154 pKa = 11.84VNTLCRR160 pKa = 11.84LALGVHH166 pKa = 6.66PGDD169 pKa = 4.17LTWVPAVTSTAIQVSRR185 pKa = 11.84TLVEE189 pKa = 4.01GTRR192 pKa = 11.84RR193 pKa = 11.84ATAQARR199 pKa = 11.84GWIRR203 pKa = 11.84AAVAVAEE210 pKa = 4.83RR211 pKa = 11.84EE212 pKa = 4.34LCPPPALEE220 pKa = 4.01EE221 pKa = 4.15LKK223 pKa = 10.62PVAKK227 pKa = 10.38GPRR230 pKa = 11.84QEE232 pKa = 4.07VGEE235 pKa = 4.05AASWVNAEE243 pKa = 4.19SVAMEE248 pKa = 4.07LRR250 pKa = 11.84ALFGLLTDD258 pKa = 4.19TPVHH262 pKa = 6.51RR263 pKa = 11.84EE264 pKa = 3.42MGGRR268 pKa = 11.84VARR271 pKa = 11.84EE272 pKa = 3.67ILRR275 pKa = 11.84DD276 pKa = 3.22RR277 pKa = 11.84CRR279 pKa = 11.84CGRR282 pKa = 11.84EE283 pKa = 3.98DD284 pKa = 3.03TWYY287 pKa = 10.81LSTSAVTHH295 pKa = 6.2WLSPTLTDD303 pKa = 4.14LVLATRR309 pKa = 11.84PQGFHH314 pKa = 6.35
Molecular weight: 33.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG29|A0A1L3KG29_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 3 OX=1922817 PE=4 SV=1
MM1 pKa = 7.63ASSTNEE7 pKa = 3.43QSGEE11 pKa = 3.68NRR13 pKa = 11.84VRR15 pKa = 11.84QGRR18 pKa = 11.84TAQRR22 pKa = 11.84PTRR25 pKa = 11.84RR26 pKa = 11.84GHH28 pKa = 5.26RR29 pKa = 11.84QGARR33 pKa = 11.84GKK35 pKa = 8.98QPRR38 pKa = 11.84GTDD41 pKa = 2.87TRR43 pKa = 11.84TSQRR47 pKa = 11.84PDD49 pKa = 3.84NIHH52 pKa = 6.45SPAPTKK58 pKa = 10.38DD59 pKa = 3.25AQQGGRR65 pKa = 11.84NTAKK69 pKa = 10.26VPAHH73 pKa = 5.69SKK75 pKa = 9.56HH76 pKa = 6.11RR77 pKa = 11.84RR78 pKa = 11.84SALHH82 pKa = 5.64RR83 pKa = 11.84ASRR86 pKa = 11.84GGIHH90 pKa = 6.06TPRR93 pKa = 11.84PRR95 pKa = 11.84QRR97 pKa = 11.84SGGSRR102 pKa = 11.84AVGSRR107 pKa = 11.84HH108 pKa = 6.48APSQPPGGYY117 pKa = 9.23QPSASQVEE125 pKa = 4.51RR126 pKa = 11.84WAKK129 pKa = 10.08IDD131 pKa = 3.62SLLPALLDD139 pKa = 3.72TIEE142 pKa = 4.1RR143 pKa = 11.84HH144 pKa = 5.15EE145 pKa = 4.33LRR147 pKa = 11.84PSEE150 pKa = 4.13VFQRR154 pKa = 11.84GLRR157 pKa = 11.84VARR160 pKa = 11.84NAVRR164 pKa = 11.84ARR166 pKa = 11.84RR167 pKa = 11.84QPVQPVPSVVTPDD180 pKa = 3.48RR181 pKa = 11.84GGEE184 pKa = 4.04PQLPAPPPTNPAGLPHH200 pKa = 7.27DD201 pKa = 5.08GEE203 pKa = 5.02GCTTGDD209 pKa = 3.48CNAAVLLSRR218 pKa = 11.84GNVKK222 pKa = 9.66PVCHH226 pKa = 5.59AHH228 pKa = 6.22KK229 pKa = 10.1WDD231 pKa = 3.58VRR233 pKa = 11.84QLQHH237 pKa = 6.94KK238 pKa = 9.5
MM1 pKa = 7.63ASSTNEE7 pKa = 3.43QSGEE11 pKa = 3.68NRR13 pKa = 11.84VRR15 pKa = 11.84QGRR18 pKa = 11.84TAQRR22 pKa = 11.84PTRR25 pKa = 11.84RR26 pKa = 11.84GHH28 pKa = 5.26RR29 pKa = 11.84QGARR33 pKa = 11.84GKK35 pKa = 8.98QPRR38 pKa = 11.84GTDD41 pKa = 2.87TRR43 pKa = 11.84TSQRR47 pKa = 11.84PDD49 pKa = 3.84NIHH52 pKa = 6.45SPAPTKK58 pKa = 10.38DD59 pKa = 3.25AQQGGRR65 pKa = 11.84NTAKK69 pKa = 10.26VPAHH73 pKa = 5.69SKK75 pKa = 9.56HH76 pKa = 6.11RR77 pKa = 11.84RR78 pKa = 11.84SALHH82 pKa = 5.64RR83 pKa = 11.84ASRR86 pKa = 11.84GGIHH90 pKa = 6.06TPRR93 pKa = 11.84PRR95 pKa = 11.84QRR97 pKa = 11.84SGGSRR102 pKa = 11.84AVGSRR107 pKa = 11.84HH108 pKa = 6.48APSQPPGGYY117 pKa = 9.23QPSASQVEE125 pKa = 4.51RR126 pKa = 11.84WAKK129 pKa = 10.08IDD131 pKa = 3.62SLLPALLDD139 pKa = 3.72TIEE142 pKa = 4.1RR143 pKa = 11.84HH144 pKa = 5.15EE145 pKa = 4.33LRR147 pKa = 11.84PSEE150 pKa = 4.13VFQRR154 pKa = 11.84GLRR157 pKa = 11.84VARR160 pKa = 11.84NAVRR164 pKa = 11.84ARR166 pKa = 11.84RR167 pKa = 11.84QPVQPVPSVVTPDD180 pKa = 3.48RR181 pKa = 11.84GGEE184 pKa = 4.04PQLPAPPPTNPAGLPHH200 pKa = 7.27DD201 pKa = 5.08GEE203 pKa = 5.02GCTTGDD209 pKa = 3.48CNAAVLLSRR218 pKa = 11.84GNVKK222 pKa = 9.66PVCHH226 pKa = 5.59AHH228 pKa = 6.22KK229 pKa = 10.1WDD231 pKa = 3.58VRR233 pKa = 11.84QLQHH237 pKa = 6.94KK238 pKa = 9.5
Molecular weight: 25.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1313 |
226 |
535 |
328.3 |
36.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.606 ± 1.654 | 1.523 ± 0.31 |
4.57 ± 0.41 | 5.179 ± 0.633 |
2.894 ± 1.142 | 6.702 ± 0.778 |
2.818 ± 0.589 | 3.046 ± 0.579 |
3.656 ± 1.325 | 8.911 ± 1.293 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.133 ± 0.676 | 3.58 ± 0.87 |
8.073 ± 1.139 | 3.503 ± 1.043 |
8.835 ± 1.443 | 7.312 ± 0.381 |
6.778 ± 0.497 | 7.845 ± 0.611 |
1.752 ± 0.366 | 2.285 ± 0.828 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
