
Coraliomargarita akajimensis (strain DSM 45221 / IAM 15411 / JCM 23193 / KCTC 12865 / 04OKA010-24)
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Opitutae; Puniceicoccales; Puniceicoccaceae; Coraliomargarita; Coraliomargarita akajimensis
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
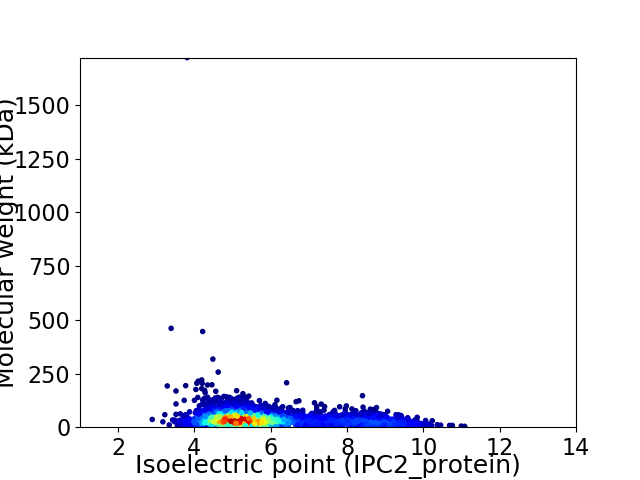
Virtual 2D-PAGE plot for 3110 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
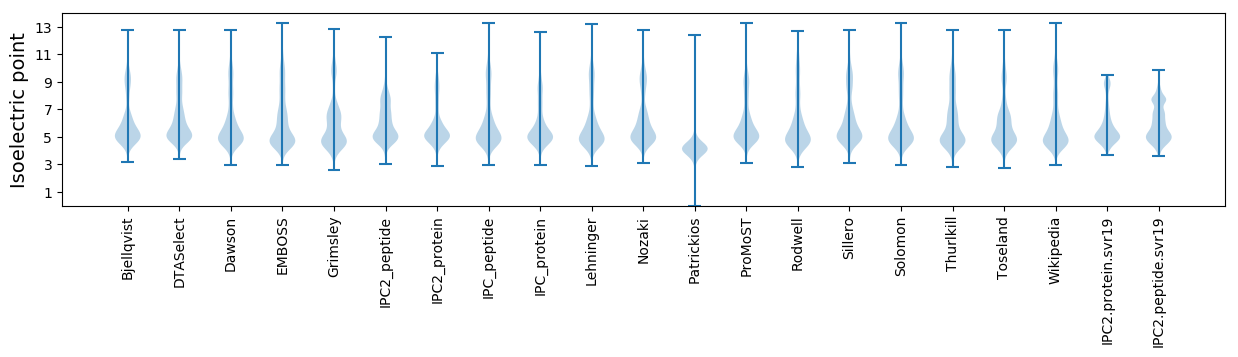
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5EMR3|D5EMR3_CORAD 3-phosphoshikimate 1-carboxyvinyltransferase OS=Coraliomargarita akajimensis (strain DSM 45221 / IAM 15411 / JCM 23193 / KCTC 12865 / 04OKA010-24) OX=583355 GN=aroA PE=3 SV=1
MM1 pKa = 7.7KK2 pKa = 10.11KK3 pKa = 8.18QTLISMFAAAATSYY17 pKa = 11.16SFAGVAGAPNGNGLYY32 pKa = 10.23EE33 pKa = 4.01ITDD36 pKa = 4.96PISADD41 pKa = 3.4TVLTSDD47 pKa = 4.15KK48 pKa = 10.7EE49 pKa = 4.21YY50 pKa = 11.09LLTEE54 pKa = 4.04IVYY57 pKa = 10.64VSDD60 pKa = 3.2ATLTIEE66 pKa = 5.07AGTTIYY72 pKa = 10.89GVVGVNGTDD81 pKa = 3.68PEE83 pKa = 4.85SGDD86 pKa = 3.92PIALVEE92 pKa = 4.62PGTLVITRR100 pKa = 11.84TAEE103 pKa = 3.38IDD105 pKa = 3.51AVGTPTSPIIFTYY118 pKa = 10.56FGDD121 pKa = 4.02ADD123 pKa = 3.8PRR125 pKa = 11.84NTPTAPLTNDD135 pKa = 3.51TLNTGLWGGVIILGNAPTNNDD156 pKa = 2.78QSADD160 pKa = 3.52NSLLVPGTDD169 pKa = 3.45AIEE172 pKa = 5.25GIVEE176 pKa = 4.31PNPADD181 pKa = 3.9LSPDD185 pKa = 2.94LRR187 pKa = 11.84GIYY190 pKa = 10.49GGDD193 pKa = 3.57APHH196 pKa = 7.44DD197 pKa = 3.9SSGVMRR203 pKa = 11.84YY204 pKa = 9.02VSIRR208 pKa = 11.84NGGFEE213 pKa = 4.41LGAANEE219 pKa = 4.35INGLTMGGVGAGTVLEE235 pKa = 4.51YY236 pKa = 10.67IEE238 pKa = 5.39VIAGQDD244 pKa = 4.4DD245 pKa = 3.85GFEE248 pKa = 4.28WFGGTVNSKK257 pKa = 10.62YY258 pKa = 10.83LLSAYY263 pKa = 10.81NNDD266 pKa = 3.73DD267 pKa = 4.23AFDD270 pKa = 4.1YY271 pKa = 11.23DD272 pKa = 3.65QGFSGLGQFWAVVQADD288 pKa = 3.68NSNDD292 pKa = 3.14GDD294 pKa = 3.84KK295 pKa = 11.31AFEE298 pKa = 4.49FDD300 pKa = 5.52GDD302 pKa = 3.95DD303 pKa = 3.33QALAKK308 pKa = 9.91PYY310 pKa = 10.05AVPVIYY316 pKa = 10.48NATIIGTGTEE326 pKa = 4.15YY327 pKa = 11.08KK328 pKa = 10.35AGNGGGQVDD337 pKa = 3.89YY338 pKa = 11.12DD339 pKa = 3.93KK340 pKa = 11.57NAGGGLFNSIIVASGIEE357 pKa = 4.06SLDD360 pKa = 4.04LNGTCIDD367 pKa = 4.12MYY369 pKa = 11.24NAGLLQIRR377 pKa = 11.84SNIWHH382 pKa = 7.38RR383 pKa = 11.84DD384 pKa = 2.96IDD386 pKa = 4.05DD387 pKa = 4.3LAAEE391 pKa = 4.35AVVDD395 pKa = 4.39HH396 pKa = 7.35DD397 pKa = 5.57AGDD400 pKa = 3.94ADD402 pKa = 4.67VPLSDD407 pKa = 3.82AANFNVFSNPFLGGANYY424 pKa = 10.96AVMGEE429 pKa = 4.06TGTAKK434 pKa = 10.04KK435 pKa = 9.8RR436 pKa = 11.84YY437 pKa = 8.28EE438 pKa = 3.93VGGVYY443 pKa = 10.6LKK445 pKa = 10.51PAVFAGSDD453 pKa = 3.62VITNLEE459 pKa = 4.19PYY461 pKa = 9.8IGTFFDD467 pKa = 3.64VVSYY471 pKa = 10.84KK472 pKa = 10.9GAFDD476 pKa = 4.57PNGTPWTAGWTVPAQLGYY494 pKa = 10.2FADD497 pKa = 3.81
MM1 pKa = 7.7KK2 pKa = 10.11KK3 pKa = 8.18QTLISMFAAAATSYY17 pKa = 11.16SFAGVAGAPNGNGLYY32 pKa = 10.23EE33 pKa = 4.01ITDD36 pKa = 4.96PISADD41 pKa = 3.4TVLTSDD47 pKa = 4.15KK48 pKa = 10.7EE49 pKa = 4.21YY50 pKa = 11.09LLTEE54 pKa = 4.04IVYY57 pKa = 10.64VSDD60 pKa = 3.2ATLTIEE66 pKa = 5.07AGTTIYY72 pKa = 10.89GVVGVNGTDD81 pKa = 3.68PEE83 pKa = 4.85SGDD86 pKa = 3.92PIALVEE92 pKa = 4.62PGTLVITRR100 pKa = 11.84TAEE103 pKa = 3.38IDD105 pKa = 3.51AVGTPTSPIIFTYY118 pKa = 10.56FGDD121 pKa = 4.02ADD123 pKa = 3.8PRR125 pKa = 11.84NTPTAPLTNDD135 pKa = 3.51TLNTGLWGGVIILGNAPTNNDD156 pKa = 2.78QSADD160 pKa = 3.52NSLLVPGTDD169 pKa = 3.45AIEE172 pKa = 5.25GIVEE176 pKa = 4.31PNPADD181 pKa = 3.9LSPDD185 pKa = 2.94LRR187 pKa = 11.84GIYY190 pKa = 10.49GGDD193 pKa = 3.57APHH196 pKa = 7.44DD197 pKa = 3.9SSGVMRR203 pKa = 11.84YY204 pKa = 9.02VSIRR208 pKa = 11.84NGGFEE213 pKa = 4.41LGAANEE219 pKa = 4.35INGLTMGGVGAGTVLEE235 pKa = 4.51YY236 pKa = 10.67IEE238 pKa = 5.39VIAGQDD244 pKa = 4.4DD245 pKa = 3.85GFEE248 pKa = 4.28WFGGTVNSKK257 pKa = 10.62YY258 pKa = 10.83LLSAYY263 pKa = 10.81NNDD266 pKa = 3.73DD267 pKa = 4.23AFDD270 pKa = 4.1YY271 pKa = 11.23DD272 pKa = 3.65QGFSGLGQFWAVVQADD288 pKa = 3.68NSNDD292 pKa = 3.14GDD294 pKa = 3.84KK295 pKa = 11.31AFEE298 pKa = 4.49FDD300 pKa = 5.52GDD302 pKa = 3.95DD303 pKa = 3.33QALAKK308 pKa = 9.91PYY310 pKa = 10.05AVPVIYY316 pKa = 10.48NATIIGTGTEE326 pKa = 4.15YY327 pKa = 11.08KK328 pKa = 10.35AGNGGGQVDD337 pKa = 3.89YY338 pKa = 11.12DD339 pKa = 3.93KK340 pKa = 11.57NAGGGLFNSIIVASGIEE357 pKa = 4.06SLDD360 pKa = 4.04LNGTCIDD367 pKa = 4.12MYY369 pKa = 11.24NAGLLQIRR377 pKa = 11.84SNIWHH382 pKa = 7.38RR383 pKa = 11.84DD384 pKa = 2.96IDD386 pKa = 4.05DD387 pKa = 4.3LAAEE391 pKa = 4.35AVVDD395 pKa = 4.39HH396 pKa = 7.35DD397 pKa = 5.57AGDD400 pKa = 3.94ADD402 pKa = 4.67VPLSDD407 pKa = 3.82AANFNVFSNPFLGGANYY424 pKa = 10.96AVMGEE429 pKa = 4.06TGTAKK434 pKa = 10.04KK435 pKa = 9.8RR436 pKa = 11.84YY437 pKa = 8.28EE438 pKa = 3.93VGGVYY443 pKa = 10.6LKK445 pKa = 10.51PAVFAGSDD453 pKa = 3.62VITNLEE459 pKa = 4.19PYY461 pKa = 9.8IGTFFDD467 pKa = 3.64VVSYY471 pKa = 10.84KK472 pKa = 10.9GAFDD476 pKa = 4.57PNGTPWTAGWTVPAQLGYY494 pKa = 10.2FADD497 pKa = 3.81
Molecular weight: 51.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5EMZ9|D5EMZ9_CORAD Putative acyl carrier protein OS=Coraliomargarita akajimensis (strain DSM 45221 / IAM 15411 / JCM 23193 / KCTC 12865 / 04OKA010-24) OX=583355 GN=Caka_2373 PE=4 SV=1
MM1 pKa = 7.59GNLKK5 pKa = 10.05KK6 pKa = 10.26KK7 pKa = 10.28RR8 pKa = 11.84RR9 pKa = 11.84LKK11 pKa = 9.46MSKK14 pKa = 9.18HH15 pKa = 5.32KK16 pKa = 10.09RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.03RR20 pKa = 11.84LKK22 pKa = 9.94SNRR25 pKa = 11.84HH26 pKa = 5.57KK27 pKa = 10.88KK28 pKa = 8.44RR29 pKa = 11.84TWW31 pKa = 2.8
MM1 pKa = 7.59GNLKK5 pKa = 10.05KK6 pKa = 10.26KK7 pKa = 10.28RR8 pKa = 11.84RR9 pKa = 11.84LKK11 pKa = 9.46MSKK14 pKa = 9.18HH15 pKa = 5.32KK16 pKa = 10.09RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.03RR20 pKa = 11.84LKK22 pKa = 9.94SNRR25 pKa = 11.84HH26 pKa = 5.57KK27 pKa = 10.88KK28 pKa = 8.44RR29 pKa = 11.84TWW31 pKa = 2.8
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1118454 |
30 |
16477 |
359.6 |
39.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.406 ± 0.048 | 1.088 ± 0.025 |
5.922 ± 0.05 | 6.643 ± 0.047 |
4.121 ± 0.03 | 7.628 ± 0.075 |
2.143 ± 0.034 | 5.662 ± 0.037 |
4.194 ± 0.064 | 9.995 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.235 ± 0.031 | 3.586 ± 0.043 |
4.5 ± 0.043 | 3.905 ± 0.031 |
5.524 ± 0.063 | 6.612 ± 0.052 |
5.506 ± 0.086 | 6.776 ± 0.041 |
1.433 ± 0.029 | 3.118 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
