
Microvirga subterranea
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Microvirga
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
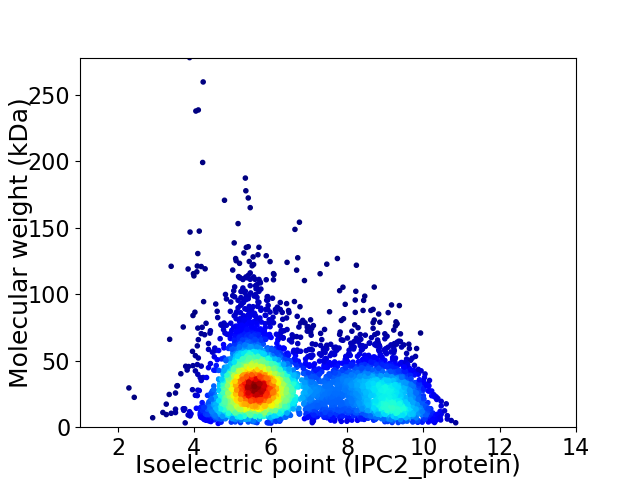
Virtual 2D-PAGE plot for 4897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370HZ70|A0A370HZ70_9RHIZ Uncharacterized protein OS=Microvirga subterranea OX=186651 GN=DES45_101512 PE=4 SV=1
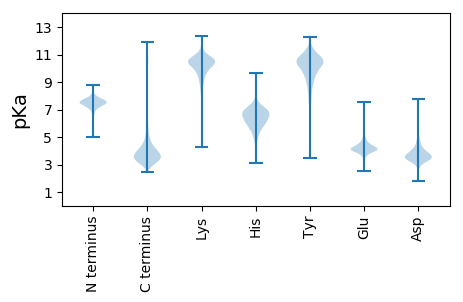
MM1 pKa = 7.82AFFDD5 pKa = 3.62VTNAFGQGFNMSATGDD21 pKa = 3.85SGWSFVEE28 pKa = 3.62ADD30 pKa = 3.96PNVSEE35 pKa = 5.02QLYY38 pKa = 10.75SDD40 pKa = 4.17DD41 pKa = 4.09GKK43 pKa = 11.41LITFSVNGSPYY54 pKa = 9.22INFYY58 pKa = 10.9GFTYY62 pKa = 9.81TLGYY66 pKa = 8.76TYY68 pKa = 10.53PDD70 pKa = 3.46GNRR73 pKa = 11.84DD74 pKa = 3.63VLIDD78 pKa = 4.25NIWYY82 pKa = 9.66YY83 pKa = 11.91NNGQNVMSVTQLNLQTTIFDD103 pKa = 3.93LQGNAWSVRR112 pKa = 11.84LNNGNDD118 pKa = 3.34TFDD121 pKa = 3.87GNDD124 pKa = 3.37YY125 pKa = 11.1NDD127 pKa = 3.78VIRR130 pKa = 11.84GGFGNDD136 pKa = 4.41LIIGYY141 pKa = 10.06AGDD144 pKa = 5.64DD145 pKa = 3.74ILLGDD150 pKa = 4.56QGNDD154 pKa = 3.56TIGGGAGNDD163 pKa = 4.17LIFGGDD169 pKa = 3.47GYY171 pKa = 10.67DD172 pKa = 3.24TVSYY176 pKa = 11.2GGFSTNYY183 pKa = 8.76IFTRR187 pKa = 11.84NVDD190 pKa = 3.38GSVTVTDD197 pKa = 3.87TTGGWGTDD205 pKa = 2.95TVYY208 pKa = 10.92DD209 pKa = 3.64VEE211 pKa = 5.43AFYY214 pKa = 11.04FNNGTFSLSSLLPIDD229 pKa = 4.79PPAPPPPPPPPPPTPEE245 pKa = 3.98EE246 pKa = 4.15IYY248 pKa = 10.86AGEE251 pKa = 4.09DD252 pKa = 3.19TLYY255 pKa = 9.54GTSGSNTLKK264 pKa = 10.87GYY266 pKa = 10.26GGNDD270 pKa = 3.41TLRR273 pKa = 11.84GQGGNDD279 pKa = 3.3YY280 pKa = 11.23LFGGDD285 pKa = 4.42GNDD288 pKa = 3.26VLYY291 pKa = 11.21GGAGKK296 pKa = 10.08DD297 pKa = 3.08AFVFDD302 pKa = 4.22TKK304 pKa = 10.88PNKK307 pKa = 7.57TTNKK311 pKa = 10.26DD312 pKa = 3.29AIKK315 pKa = 10.33DD316 pKa = 3.87FKK318 pKa = 11.56VVDD321 pKa = 3.34DD322 pKa = 5.12TIRR325 pKa = 11.84LDD327 pKa = 3.33NAVFTKK333 pKa = 10.82VGGNGTLKK341 pKa = 10.91ASAFWTNTTGKK352 pKa = 10.1AHH354 pKa = 7.21DD355 pKa = 3.83KK356 pKa = 10.21DD357 pKa = 3.99DD358 pKa = 3.73RR359 pKa = 11.84VIYY362 pKa = 10.75DD363 pKa = 3.29KK364 pKa = 11.41DD365 pKa = 3.89SGVLYY370 pKa = 10.78YY371 pKa = 10.79DD372 pKa = 4.17ADD374 pKa = 4.04GSGKK378 pKa = 9.86GAAVAFATISKK389 pKa = 10.18NLALTNKK396 pKa = 9.91DD397 pKa = 3.61FYY399 pKa = 11.0IVV401 pKa = 3.15
MM1 pKa = 7.82AFFDD5 pKa = 3.62VTNAFGQGFNMSATGDD21 pKa = 3.85SGWSFVEE28 pKa = 3.62ADD30 pKa = 3.96PNVSEE35 pKa = 5.02QLYY38 pKa = 10.75SDD40 pKa = 4.17DD41 pKa = 4.09GKK43 pKa = 11.41LITFSVNGSPYY54 pKa = 9.22INFYY58 pKa = 10.9GFTYY62 pKa = 9.81TLGYY66 pKa = 8.76TYY68 pKa = 10.53PDD70 pKa = 3.46GNRR73 pKa = 11.84DD74 pKa = 3.63VLIDD78 pKa = 4.25NIWYY82 pKa = 9.66YY83 pKa = 11.91NNGQNVMSVTQLNLQTTIFDD103 pKa = 3.93LQGNAWSVRR112 pKa = 11.84LNNGNDD118 pKa = 3.34TFDD121 pKa = 3.87GNDD124 pKa = 3.37YY125 pKa = 11.1NDD127 pKa = 3.78VIRR130 pKa = 11.84GGFGNDD136 pKa = 4.41LIIGYY141 pKa = 10.06AGDD144 pKa = 5.64DD145 pKa = 3.74ILLGDD150 pKa = 4.56QGNDD154 pKa = 3.56TIGGGAGNDD163 pKa = 4.17LIFGGDD169 pKa = 3.47GYY171 pKa = 10.67DD172 pKa = 3.24TVSYY176 pKa = 11.2GGFSTNYY183 pKa = 8.76IFTRR187 pKa = 11.84NVDD190 pKa = 3.38GSVTVTDD197 pKa = 3.87TTGGWGTDD205 pKa = 2.95TVYY208 pKa = 10.92DD209 pKa = 3.64VEE211 pKa = 5.43AFYY214 pKa = 11.04FNNGTFSLSSLLPIDD229 pKa = 4.79PPAPPPPPPPPPPTPEE245 pKa = 3.98EE246 pKa = 4.15IYY248 pKa = 10.86AGEE251 pKa = 4.09DD252 pKa = 3.19TLYY255 pKa = 9.54GTSGSNTLKK264 pKa = 10.87GYY266 pKa = 10.26GGNDD270 pKa = 3.41TLRR273 pKa = 11.84GQGGNDD279 pKa = 3.3YY280 pKa = 11.23LFGGDD285 pKa = 4.42GNDD288 pKa = 3.26VLYY291 pKa = 11.21GGAGKK296 pKa = 10.08DD297 pKa = 3.08AFVFDD302 pKa = 4.22TKK304 pKa = 10.88PNKK307 pKa = 7.57TTNKK311 pKa = 10.26DD312 pKa = 3.29AIKK315 pKa = 10.33DD316 pKa = 3.87FKK318 pKa = 11.56VVDD321 pKa = 3.34DD322 pKa = 5.12TIRR325 pKa = 11.84LDD327 pKa = 3.33NAVFTKK333 pKa = 10.82VGGNGTLKK341 pKa = 10.91ASAFWTNTTGKK352 pKa = 10.1AHH354 pKa = 7.21DD355 pKa = 3.83KK356 pKa = 10.21DD357 pKa = 3.99DD358 pKa = 3.73RR359 pKa = 11.84VIYY362 pKa = 10.75DD363 pKa = 3.29KK364 pKa = 11.41DD365 pKa = 3.89SGVLYY370 pKa = 10.78YY371 pKa = 10.79DD372 pKa = 4.17ADD374 pKa = 4.04GSGKK378 pKa = 9.86GAAVAFATISKK389 pKa = 10.18NLALTNKK396 pKa = 9.91DD397 pKa = 3.61FYY399 pKa = 11.0IVV401 pKa = 3.15
Molecular weight: 42.97 kDa
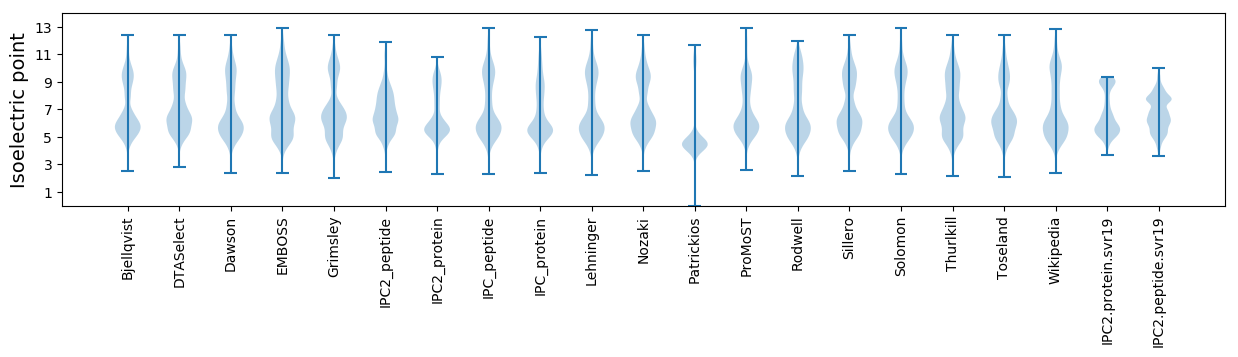
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370HXV8|A0A370HXV8_9RHIZ Xanthine dehydrogenase YagT iron-sulfur-binding subunit OS=Microvirga subterranea OX=186651 GN=DES45_10129 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.27GLPKK6 pKa = 10.1GWRR9 pKa = 11.84IHH11 pKa = 7.18DD12 pKa = 3.67GRR14 pKa = 11.84DD15 pKa = 3.3PEE17 pKa = 4.25EE18 pKa = 4.07EE19 pKa = 3.81LRR21 pKa = 11.84KK22 pKa = 9.95LKK24 pKa = 10.63RR25 pKa = 11.84RR26 pKa = 11.84FQAVSDD32 pKa = 3.65RR33 pKa = 11.84FHH35 pKa = 6.84RR36 pKa = 11.84ARR38 pKa = 11.84RR39 pKa = 11.84LRR41 pKa = 11.84SLLRR45 pKa = 11.84QIRR48 pKa = 11.84LPALLLTGIFAVVTVLVTLSPWPFTTTVRR77 pKa = 11.84HH78 pKa = 5.61LASFPSCGLARR89 pKa = 11.84AIGLAPAYY97 pKa = 10.3RR98 pKa = 11.84GDD100 pKa = 3.63PGYY103 pKa = 9.44WAHH106 pKa = 6.7QDD108 pKa = 3.2EE109 pKa = 5.05DD110 pKa = 4.29SDD112 pKa = 4.44GRR114 pKa = 11.84SCEE117 pKa = 3.92TWKK120 pKa = 10.69SHH122 pKa = 6.05
MM1 pKa = 7.54KK2 pKa = 10.27GLPKK6 pKa = 10.1GWRR9 pKa = 11.84IHH11 pKa = 7.18DD12 pKa = 3.67GRR14 pKa = 11.84DD15 pKa = 3.3PEE17 pKa = 4.25EE18 pKa = 4.07EE19 pKa = 3.81LRR21 pKa = 11.84KK22 pKa = 9.95LKK24 pKa = 10.63RR25 pKa = 11.84RR26 pKa = 11.84FQAVSDD32 pKa = 3.65RR33 pKa = 11.84FHH35 pKa = 6.84RR36 pKa = 11.84ARR38 pKa = 11.84RR39 pKa = 11.84LRR41 pKa = 11.84SLLRR45 pKa = 11.84QIRR48 pKa = 11.84LPALLLTGIFAVVTVLVTLSPWPFTTTVRR77 pKa = 11.84HH78 pKa = 5.61LASFPSCGLARR89 pKa = 11.84AIGLAPAYY97 pKa = 10.3RR98 pKa = 11.84GDD100 pKa = 3.63PGYY103 pKa = 9.44WAHH106 pKa = 6.7QDD108 pKa = 3.2EE109 pKa = 5.05DD110 pKa = 4.29SDD112 pKa = 4.44GRR114 pKa = 11.84SCEE117 pKa = 3.92TWKK120 pKa = 10.69SHH122 pKa = 6.05
Molecular weight: 13.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1507765 |
25 |
2599 |
307.9 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.033 ± 0.049 | 0.758 ± 0.011 |
5.652 ± 0.036 | 5.818 ± 0.036 |
3.704 ± 0.023 | 8.662 ± 0.041 |
2.013 ± 0.017 | 5.111 ± 0.024 |
3.198 ± 0.029 | 10.196 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.02 | 2.58 ± 0.029 |
5.342 ± 0.029 | 3.179 ± 0.022 |
7.385 ± 0.041 | 5.474 ± 0.024 |
5.333 ± 0.029 | 7.683 ± 0.029 |
1.309 ± 0.015 | 2.214 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
