
Candidatus Paracaedibacter acanthamoebae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Holosporales; Candidatus Paracaedibacteraceae; Candidatus Paracaedibacter
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
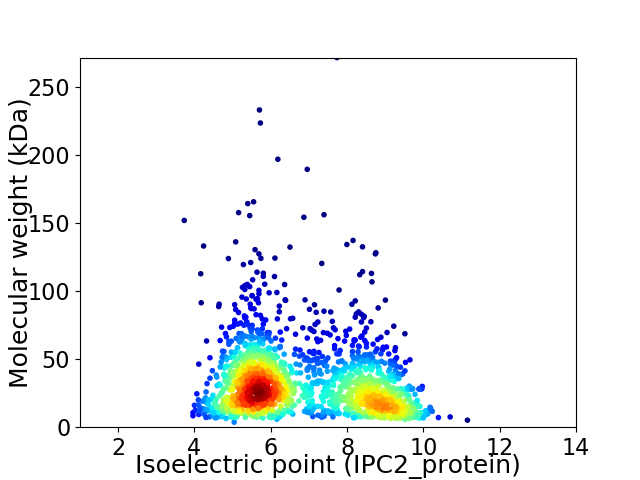
Virtual 2D-PAGE plot for 1803 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077ATH3|A0A077ATH3_9PROT Uncharacterized protein OS=Candidatus Paracaedibacter acanthamoebae OX=91604 GN=ID47_06520 PE=4 SV=1
MM1 pKa = 7.52TLQTLQTTLSQLNSTNISEE20 pKa = 4.58AVISNAGLTPRR31 pKa = 11.84EE32 pKa = 4.19DD33 pKa = 4.42LDD35 pKa = 4.46NLLQSALMLTQPLNVNIVNVGTADD59 pKa = 3.6DD60 pKa = 3.96TSLVIQATGSFLNLSSINLQIEE82 pKa = 4.3FVLNQDD88 pKa = 2.91ITDD91 pKa = 4.01VIINATLPDD100 pKa = 3.76NQGQAWQFADD110 pKa = 3.57SFPYY114 pKa = 8.89MVIYY118 pKa = 9.77PFYY121 pKa = 10.16NISFNSPSFIFSSYY135 pKa = 8.34GQPYY139 pKa = 8.0TWQSQPLSLTAGLNFASFLPLDD161 pKa = 3.82SGFVGLVSNFFSTPPSGEE179 pKa = 3.73ILFSGSVDD187 pKa = 3.1TSVIGKK193 pKa = 9.35NNIVWPSVNFSGLMSDD209 pKa = 3.85QPSDD213 pKa = 4.24FIPGLEE219 pKa = 4.02FTNPIITLTSQIDD232 pKa = 3.95LDD234 pKa = 4.56DD235 pKa = 4.01MQSYY239 pKa = 10.93SLNFSVTEE247 pKa = 4.0DD248 pKa = 3.43QIGLNLEE255 pKa = 4.15TQLQNNAVVGFSIVPSDD272 pKa = 3.58STNPPSLNTVVQLIPGLTAADD293 pKa = 3.59ITNDD297 pKa = 3.09IPQEE301 pKa = 3.99LQQSFSAITFVEE313 pKa = 5.06ASCTLCINPINILSMTLVLSAGSAWAITDD342 pKa = 3.43EE343 pKa = 4.45FTVKK347 pKa = 10.46SLTLSYY353 pKa = 10.49TVLDD357 pKa = 4.05PLGTPTNIFGFNGVLSFYY375 pKa = 10.73PEE377 pKa = 4.0VFKK380 pKa = 11.29GLFDD384 pKa = 3.53IQISIDD390 pKa = 3.51TTNNNLLVSGSYY402 pKa = 9.98QGIIDD407 pKa = 5.18LNTITTNLCNLSLPSQFSSINVTDD431 pKa = 4.1FVVSFNKK438 pKa = 9.96INTNWTWGLFCNVDD452 pKa = 3.32GTFPLPFVQGSVDD465 pKa = 3.9CNLSASLSSNSYY477 pKa = 9.02YY478 pKa = 10.93LEE480 pKa = 4.74GGLTIGGSYY489 pKa = 10.73FNVSFDD495 pKa = 4.3SNLKK499 pKa = 9.67NSVLAGYY506 pKa = 7.65WKK508 pKa = 10.6SLSSDD513 pKa = 3.76DD514 pKa = 4.53LLGIQSLADD523 pKa = 3.25AVGIKK528 pKa = 8.42VTIPTTFDD536 pKa = 3.86LDD538 pKa = 3.46LTQAGFSYY546 pKa = 11.26NLTQNTVKK554 pKa = 11.03LEE556 pKa = 4.07ATSLNYY562 pKa = 10.72GKK564 pKa = 9.2GTFATFLPKK573 pKa = 9.65GTADD577 pKa = 2.91QYY579 pKa = 11.78EE580 pKa = 4.96FYY582 pKa = 11.04FNLDD586 pKa = 3.24VQTDD590 pKa = 3.77IVILSQDD597 pKa = 3.55LPLIGQEE604 pKa = 4.17FSTADD609 pKa = 3.79EE610 pKa = 4.4IALKK614 pKa = 10.51SLNITYY620 pKa = 10.74ASASLPNVNPIKK632 pKa = 10.67NFNSGLDD639 pKa = 3.34IKK641 pKa = 10.99GVLSIAGDD649 pKa = 3.74DD650 pKa = 4.13HH651 pKa = 7.53PFDD654 pKa = 5.73LDD656 pKa = 4.01FGPAVQTEE664 pKa = 4.48SGGAPSLITDD674 pKa = 4.34LTATSSTPPPTPPGVWVNIQKK695 pKa = 8.61TVGPISLKK703 pKa = 10.62RR704 pKa = 11.84IGLNYY709 pKa = 10.19QNDD712 pKa = 4.45LMWIMLDD719 pKa = 3.54ASLNTTNLNIALTGLSVGMNVSNSEE744 pKa = 3.95PDD746 pKa = 3.44LQFALHH752 pKa = 6.6GLDD755 pKa = 4.27VDD757 pKa = 4.42LTSDD761 pKa = 4.72DD762 pKa = 3.4IVINGGLLSADD773 pKa = 3.44DD774 pKa = 4.13SGGTEE779 pKa = 3.98YY780 pKa = 11.11DD781 pKa = 3.04GTLTVKK787 pKa = 10.65VGEE790 pKa = 4.1FDD792 pKa = 3.22IAALASYY799 pKa = 10.85AMPPDD804 pKa = 4.18DD805 pKa = 4.64QSSLFAFAVIDD816 pKa = 4.18FPLGGPPAFFITGGAGGFGYY836 pKa = 10.6NRR838 pKa = 11.84DD839 pKa = 3.83LIMPNISSIMSFPLIEE855 pKa = 4.24IAQNPSKK862 pKa = 10.99YY863 pKa = 10.15KK864 pKa = 10.73NEE866 pKa = 4.02NLSSLLQTVQSAIPIEE882 pKa = 4.18QGQYY886 pKa = 8.65WLAAGINFNSYY897 pKa = 10.87GMVNSYY903 pKa = 10.9LLAAAAFGTQLEE915 pKa = 4.42LDD917 pKa = 4.08LLGVSTIVAPPLDD930 pKa = 4.79PDD932 pKa = 4.09PVAMIQFAIEE942 pKa = 4.16TTLVPAEE949 pKa = 4.16GLFSVEE955 pKa = 4.08GQLTNNSYY963 pKa = 10.98VLSKK967 pKa = 10.7NCHH970 pKa = 4.75LTGGFAFYY978 pKa = 10.6LWFGSNPHH986 pKa = 6.81AGDD989 pKa = 3.73FVISLGGYY997 pKa = 7.62SPKK1000 pKa = 10.16FKK1002 pKa = 10.62KK1003 pKa = 9.78PSYY1006 pKa = 9.59YY1007 pKa = 9.06PTVPLLGLNWQVCPEE1022 pKa = 4.36LSIQGGLYY1030 pKa = 10.37CAITSLAIMVGGSLKK1045 pKa = 10.77ALWQSDD1051 pKa = 4.19GIRR1054 pKa = 11.84AWFNLDD1060 pKa = 3.32ADD1062 pKa = 4.66VLLFWQPYY1070 pKa = 9.61QYY1072 pKa = 10.78TIDD1075 pKa = 3.75ANVDD1079 pKa = 3.17VGASLIIDD1087 pKa = 3.47VAFVNTAITAHH1098 pKa = 6.09TNADD1102 pKa = 3.62LSIWGPDD1109 pKa = 3.32FTGKK1113 pKa = 10.6AKK1115 pKa = 10.44VDD1117 pKa = 3.64LYY1119 pKa = 10.54IISFTISFGDD1129 pKa = 3.79SEE1131 pKa = 5.74PITTPPAAISWSNFKK1146 pKa = 10.71QSFLSTDD1153 pKa = 3.32PSNVCNLNLAHH1164 pKa = 6.79GVIKK1168 pKa = 10.7SDD1170 pKa = 4.34VSDD1173 pKa = 3.93QGITYY1178 pKa = 10.2DD1179 pKa = 3.61WIVDD1183 pKa = 3.55PEE1185 pKa = 4.28HH1186 pKa = 7.11FIFTTGSSIPSKK1198 pKa = 9.41TLSLNGVAQTLSTMDD1213 pKa = 3.33SSINTDD1219 pKa = 3.73FGVGMVRR1226 pKa = 11.84APSDD1230 pKa = 3.56SFTSDD1235 pKa = 2.69HH1236 pKa = 7.16AITLTYY1242 pKa = 10.64LDD1244 pKa = 4.15ADD1246 pKa = 3.71GNEE1249 pKa = 4.7PYY1251 pKa = 10.64DD1252 pKa = 3.61IADD1255 pKa = 3.9KK1256 pKa = 11.14FNISLLTSNVPQSLWQCITSLDD1278 pKa = 3.87DD1279 pKa = 4.52LEE1281 pKa = 4.62TSVASSSTTTIANTLSGISFSAKK1304 pKa = 8.26VTPADD1309 pKa = 3.28QVTPVALHH1317 pKa = 6.05NLLYY1321 pKa = 10.92SIDD1324 pKa = 3.97FEE1326 pKa = 4.53EE1327 pKa = 5.98DD1328 pKa = 2.92NLISYY1333 pKa = 7.75NKK1335 pKa = 9.57PSVPTTDD1342 pKa = 3.53GFTEE1346 pKa = 4.73EE1347 pKa = 4.21PCAEE1351 pKa = 4.06IGQTIAKK1358 pKa = 6.77NTSRR1362 pKa = 11.84TSILTRR1368 pKa = 11.84LQTTSFKK1375 pKa = 10.45IINDD1379 pKa = 3.32INVTSLSTEE1388 pKa = 3.58AANVYY1393 pKa = 9.98FVGDD1397 pKa = 4.71PILSYY1402 pKa = 10.86LGEE1405 pKa = 4.15EE1406 pKa = 4.3KK1407 pKa = 11.05VLGGASS1413 pKa = 3.13
MM1 pKa = 7.52TLQTLQTTLSQLNSTNISEE20 pKa = 4.58AVISNAGLTPRR31 pKa = 11.84EE32 pKa = 4.19DD33 pKa = 4.42LDD35 pKa = 4.46NLLQSALMLTQPLNVNIVNVGTADD59 pKa = 3.6DD60 pKa = 3.96TSLVIQATGSFLNLSSINLQIEE82 pKa = 4.3FVLNQDD88 pKa = 2.91ITDD91 pKa = 4.01VIINATLPDD100 pKa = 3.76NQGQAWQFADD110 pKa = 3.57SFPYY114 pKa = 8.89MVIYY118 pKa = 9.77PFYY121 pKa = 10.16NISFNSPSFIFSSYY135 pKa = 8.34GQPYY139 pKa = 8.0TWQSQPLSLTAGLNFASFLPLDD161 pKa = 3.82SGFVGLVSNFFSTPPSGEE179 pKa = 3.73ILFSGSVDD187 pKa = 3.1TSVIGKK193 pKa = 9.35NNIVWPSVNFSGLMSDD209 pKa = 3.85QPSDD213 pKa = 4.24FIPGLEE219 pKa = 4.02FTNPIITLTSQIDD232 pKa = 3.95LDD234 pKa = 4.56DD235 pKa = 4.01MQSYY239 pKa = 10.93SLNFSVTEE247 pKa = 4.0DD248 pKa = 3.43QIGLNLEE255 pKa = 4.15TQLQNNAVVGFSIVPSDD272 pKa = 3.58STNPPSLNTVVQLIPGLTAADD293 pKa = 3.59ITNDD297 pKa = 3.09IPQEE301 pKa = 3.99LQQSFSAITFVEE313 pKa = 5.06ASCTLCINPINILSMTLVLSAGSAWAITDD342 pKa = 3.43EE343 pKa = 4.45FTVKK347 pKa = 10.46SLTLSYY353 pKa = 10.49TVLDD357 pKa = 4.05PLGTPTNIFGFNGVLSFYY375 pKa = 10.73PEE377 pKa = 4.0VFKK380 pKa = 11.29GLFDD384 pKa = 3.53IQISIDD390 pKa = 3.51TTNNNLLVSGSYY402 pKa = 9.98QGIIDD407 pKa = 5.18LNTITTNLCNLSLPSQFSSINVTDD431 pKa = 4.1FVVSFNKK438 pKa = 9.96INTNWTWGLFCNVDD452 pKa = 3.32GTFPLPFVQGSVDD465 pKa = 3.9CNLSASLSSNSYY477 pKa = 9.02YY478 pKa = 10.93LEE480 pKa = 4.74GGLTIGGSYY489 pKa = 10.73FNVSFDD495 pKa = 4.3SNLKK499 pKa = 9.67NSVLAGYY506 pKa = 7.65WKK508 pKa = 10.6SLSSDD513 pKa = 3.76DD514 pKa = 4.53LLGIQSLADD523 pKa = 3.25AVGIKK528 pKa = 8.42VTIPTTFDD536 pKa = 3.86LDD538 pKa = 3.46LTQAGFSYY546 pKa = 11.26NLTQNTVKK554 pKa = 11.03LEE556 pKa = 4.07ATSLNYY562 pKa = 10.72GKK564 pKa = 9.2GTFATFLPKK573 pKa = 9.65GTADD577 pKa = 2.91QYY579 pKa = 11.78EE580 pKa = 4.96FYY582 pKa = 11.04FNLDD586 pKa = 3.24VQTDD590 pKa = 3.77IVILSQDD597 pKa = 3.55LPLIGQEE604 pKa = 4.17FSTADD609 pKa = 3.79EE610 pKa = 4.4IALKK614 pKa = 10.51SLNITYY620 pKa = 10.74ASASLPNVNPIKK632 pKa = 10.67NFNSGLDD639 pKa = 3.34IKK641 pKa = 10.99GVLSIAGDD649 pKa = 3.74DD650 pKa = 4.13HH651 pKa = 7.53PFDD654 pKa = 5.73LDD656 pKa = 4.01FGPAVQTEE664 pKa = 4.48SGGAPSLITDD674 pKa = 4.34LTATSSTPPPTPPGVWVNIQKK695 pKa = 8.61TVGPISLKK703 pKa = 10.62RR704 pKa = 11.84IGLNYY709 pKa = 10.19QNDD712 pKa = 4.45LMWIMLDD719 pKa = 3.54ASLNTTNLNIALTGLSVGMNVSNSEE744 pKa = 3.95PDD746 pKa = 3.44LQFALHH752 pKa = 6.6GLDD755 pKa = 4.27VDD757 pKa = 4.42LTSDD761 pKa = 4.72DD762 pKa = 3.4IVINGGLLSADD773 pKa = 3.44DD774 pKa = 4.13SGGTEE779 pKa = 3.98YY780 pKa = 11.11DD781 pKa = 3.04GTLTVKK787 pKa = 10.65VGEE790 pKa = 4.1FDD792 pKa = 3.22IAALASYY799 pKa = 10.85AMPPDD804 pKa = 4.18DD805 pKa = 4.64QSSLFAFAVIDD816 pKa = 4.18FPLGGPPAFFITGGAGGFGYY836 pKa = 10.6NRR838 pKa = 11.84DD839 pKa = 3.83LIMPNISSIMSFPLIEE855 pKa = 4.24IAQNPSKK862 pKa = 10.99YY863 pKa = 10.15KK864 pKa = 10.73NEE866 pKa = 4.02NLSSLLQTVQSAIPIEE882 pKa = 4.18QGQYY886 pKa = 8.65WLAAGINFNSYY897 pKa = 10.87GMVNSYY903 pKa = 10.9LLAAAAFGTQLEE915 pKa = 4.42LDD917 pKa = 4.08LLGVSTIVAPPLDD930 pKa = 4.79PDD932 pKa = 4.09PVAMIQFAIEE942 pKa = 4.16TTLVPAEE949 pKa = 4.16GLFSVEE955 pKa = 4.08GQLTNNSYY963 pKa = 10.98VLSKK967 pKa = 10.7NCHH970 pKa = 4.75LTGGFAFYY978 pKa = 10.6LWFGSNPHH986 pKa = 6.81AGDD989 pKa = 3.73FVISLGGYY997 pKa = 7.62SPKK1000 pKa = 10.16FKK1002 pKa = 10.62KK1003 pKa = 9.78PSYY1006 pKa = 9.59YY1007 pKa = 9.06PTVPLLGLNWQVCPEE1022 pKa = 4.36LSIQGGLYY1030 pKa = 10.37CAITSLAIMVGGSLKK1045 pKa = 10.77ALWQSDD1051 pKa = 4.19GIRR1054 pKa = 11.84AWFNLDD1060 pKa = 3.32ADD1062 pKa = 4.66VLLFWQPYY1070 pKa = 9.61QYY1072 pKa = 10.78TIDD1075 pKa = 3.75ANVDD1079 pKa = 3.17VGASLIIDD1087 pKa = 3.47VAFVNTAITAHH1098 pKa = 6.09TNADD1102 pKa = 3.62LSIWGPDD1109 pKa = 3.32FTGKK1113 pKa = 10.6AKK1115 pKa = 10.44VDD1117 pKa = 3.64LYY1119 pKa = 10.54IISFTISFGDD1129 pKa = 3.79SEE1131 pKa = 5.74PITTPPAAISWSNFKK1146 pKa = 10.71QSFLSTDD1153 pKa = 3.32PSNVCNLNLAHH1164 pKa = 6.79GVIKK1168 pKa = 10.7SDD1170 pKa = 4.34VSDD1173 pKa = 3.93QGITYY1178 pKa = 10.2DD1179 pKa = 3.61WIVDD1183 pKa = 3.55PEE1185 pKa = 4.28HH1186 pKa = 7.11FIFTTGSSIPSKK1198 pKa = 9.41TLSLNGVAQTLSTMDD1213 pKa = 3.33SSINTDD1219 pKa = 3.73FGVGMVRR1226 pKa = 11.84APSDD1230 pKa = 3.56SFTSDD1235 pKa = 2.69HH1236 pKa = 7.16AITLTYY1242 pKa = 10.64LDD1244 pKa = 4.15ADD1246 pKa = 3.71GNEE1249 pKa = 4.7PYY1251 pKa = 10.64DD1252 pKa = 3.61IADD1255 pKa = 3.9KK1256 pKa = 11.14FNISLLTSNVPQSLWQCITSLDD1278 pKa = 3.87DD1279 pKa = 4.52LEE1281 pKa = 4.62TSVASSSTTTIANTLSGISFSAKK1304 pKa = 8.26VTPADD1309 pKa = 3.28QVTPVALHH1317 pKa = 6.05NLLYY1321 pKa = 10.92SIDD1324 pKa = 3.97FEE1326 pKa = 4.53EE1327 pKa = 5.98DD1328 pKa = 2.92NLISYY1333 pKa = 7.75NKK1335 pKa = 9.57PSVPTTDD1342 pKa = 3.53GFTEE1346 pKa = 4.73EE1347 pKa = 4.21PCAEE1351 pKa = 4.06IGQTIAKK1358 pKa = 6.77NTSRR1362 pKa = 11.84TSILTRR1368 pKa = 11.84LQTTSFKK1375 pKa = 10.45IINDD1379 pKa = 3.32INVTSLSTEE1388 pKa = 3.58AANVYY1393 pKa = 9.98FVGDD1397 pKa = 4.71PILSYY1402 pKa = 10.86LGEE1405 pKa = 4.15EE1406 pKa = 4.3KK1407 pKa = 11.05VLGGASS1413 pKa = 3.13
Molecular weight: 151.79 kDa
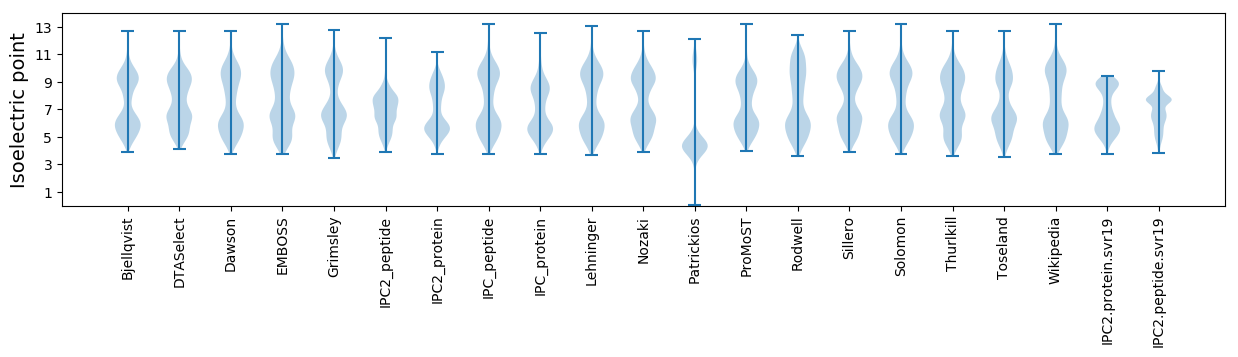
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077AYA7|A0A077AYA7_9PROT Holliday junction ATP-dependent DNA helicase RuvB OS=Candidatus Paracaedibacter acanthamoebae OX=91604 GN=ruvB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
547837 |
31 |
2490 |
303.8 |
34.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.805 ± 0.074 | 1.018 ± 0.019 |
5.146 ± 0.048 | 6.104 ± 0.065 |
4.251 ± 0.045 | 6.465 ± 0.071 |
2.369 ± 0.029 | 7.223 ± 0.046 |
6.354 ± 0.06 | 10.485 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.428 ± 0.029 | 4.355 ± 0.047 |
4.216 ± 0.043 | 4.378 ± 0.055 |
4.729 ± 0.047 | 6.753 ± 0.059 |
5.689 ± 0.044 | 5.89 ± 0.053 |
1.103 ± 0.022 | 3.237 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
