
Maribellus luteus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Maribellus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
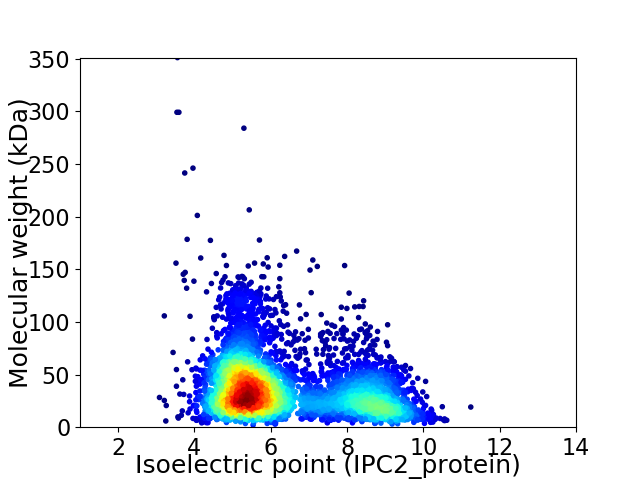
Virtual 2D-PAGE plot for 4999 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A399T8F4|A0A399T8F4_9BACT Uncharacterized protein OS=Maribellus luteus OX=2305463 GN=D1614_00420 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 10.29KK3 pKa = 10.19QIFNLFYY10 pKa = 11.15ALALLAIVTACSDD23 pKa = 3.82DD24 pKa = 4.86DD25 pKa = 5.17DD26 pKa = 6.47PIDD29 pKa = 4.12LPVYY33 pKa = 10.67GDD35 pKa = 3.33ATMTSFGFYY44 pKa = 10.65VEE46 pKa = 5.24DD47 pKa = 3.72NEE49 pKa = 5.0GVILKK54 pKa = 10.29DD55 pKa = 3.67YY56 pKa = 9.68VAEE59 pKa = 5.29GITGDD64 pKa = 3.81FTVSVPDD71 pKa = 3.88YY72 pKa = 10.49VNKK75 pKa = 8.83TALVARR81 pKa = 11.84FVVSEE86 pKa = 3.95NDD88 pKa = 3.29TVYY91 pKa = 11.25VNNVQQEE98 pKa = 4.53SGVTANDD105 pKa = 3.85FSAPLDD111 pKa = 3.96YY112 pKa = 10.6IVSEE116 pKa = 4.28GTNNTKK122 pKa = 9.19YY123 pKa = 9.27TVTVANLPAAVWSLAASDD141 pKa = 3.8TTDD144 pKa = 3.03LRR146 pKa = 11.84EE147 pKa = 3.62FSMRR151 pKa = 11.84VNPVTNVPYY160 pKa = 10.38LAYY163 pKa = 10.28ALDD166 pKa = 4.15MDD168 pKa = 5.15DD169 pKa = 5.1SADD172 pKa = 3.57EE173 pKa = 4.06KK174 pKa = 11.53VGVFKK179 pKa = 11.19LDD181 pKa = 3.09GNTLVNVGAKK191 pKa = 8.58TVSEE195 pKa = 4.42GRR197 pKa = 11.84ASYY200 pKa = 10.08PRR202 pKa = 11.84IAFDD206 pKa = 4.2AEE208 pKa = 3.95GAPFISYY215 pKa = 11.16ADD217 pKa = 3.63YY218 pKa = 11.18TNNDD222 pKa = 3.68PYY224 pKa = 11.41KK225 pKa = 10.39EE226 pKa = 4.15GSTTYY231 pKa = 10.42AASVMNYY238 pKa = 9.92NGSSWSYY245 pKa = 10.66VGNKK249 pKa = 9.83GITDD253 pKa = 3.32VRR255 pKa = 11.84ITYY258 pKa = 10.78NDD260 pKa = 3.08IVLKK264 pKa = 10.93NDD266 pKa = 3.53GNPMLFCYY274 pKa = 10.61NNAAGTLARR283 pKa = 11.84RR284 pKa = 11.84EE285 pKa = 4.12LNISDD290 pKa = 3.46FTGNSWASNLTIPGRR305 pKa = 11.84ASTQYY310 pKa = 11.18AYY312 pKa = 8.73HH313 pKa = 5.63TTAKK317 pKa = 10.42NVDD320 pKa = 3.1GVIYY324 pKa = 10.51LGVYY328 pKa = 8.63NANEE332 pKa = 4.38GTFSVYY338 pKa = 10.94KK339 pKa = 9.71NTDD342 pKa = 3.36GSWTTIVEE350 pKa = 4.47SYY352 pKa = 10.42LQDD355 pKa = 3.34GATTGNLRR363 pKa = 11.84DD364 pKa = 3.51FDD366 pKa = 4.1MDD368 pKa = 3.0VDD370 pKa = 3.79RR371 pKa = 11.84DD372 pKa = 3.99GNVFVCVADD381 pKa = 4.45DD382 pKa = 3.47EE383 pKa = 5.12AGADD387 pKa = 3.15IYY389 pKa = 10.89RR390 pKa = 11.84PKK392 pKa = 9.91VLKK395 pKa = 10.7YY396 pKa = 10.57DD397 pKa = 3.54ATAEE401 pKa = 4.18TWSKK405 pKa = 11.0VGTPIAIDD413 pKa = 4.07FSTTRR418 pKa = 11.84EE419 pKa = 3.64FSLAISPVGTPFLMYY434 pKa = 9.9RR435 pKa = 11.84DD436 pKa = 4.31DD437 pKa = 4.49SMFPVVVSFDD447 pKa = 4.36EE448 pKa = 4.57DD449 pKa = 3.89TQDD452 pKa = 2.75WTTPAQLNAVEE463 pKa = 4.81AEE465 pKa = 4.02NLYY468 pKa = 10.07MDD470 pKa = 4.65FAPNGVAYY478 pKa = 10.03AAFTTSATGEE488 pKa = 4.29TMLFKK493 pKa = 10.91YY494 pKa = 9.83DD495 pKa = 3.46IPAEE499 pKa = 4.05
MM1 pKa = 7.64KK2 pKa = 10.29KK3 pKa = 10.19QIFNLFYY10 pKa = 11.15ALALLAIVTACSDD23 pKa = 3.82DD24 pKa = 4.86DD25 pKa = 5.17DD26 pKa = 6.47PIDD29 pKa = 4.12LPVYY33 pKa = 10.67GDD35 pKa = 3.33ATMTSFGFYY44 pKa = 10.65VEE46 pKa = 5.24DD47 pKa = 3.72NEE49 pKa = 5.0GVILKK54 pKa = 10.29DD55 pKa = 3.67YY56 pKa = 9.68VAEE59 pKa = 5.29GITGDD64 pKa = 3.81FTVSVPDD71 pKa = 3.88YY72 pKa = 10.49VNKK75 pKa = 8.83TALVARR81 pKa = 11.84FVVSEE86 pKa = 3.95NDD88 pKa = 3.29TVYY91 pKa = 11.25VNNVQQEE98 pKa = 4.53SGVTANDD105 pKa = 3.85FSAPLDD111 pKa = 3.96YY112 pKa = 10.6IVSEE116 pKa = 4.28GTNNTKK122 pKa = 9.19YY123 pKa = 9.27TVTVANLPAAVWSLAASDD141 pKa = 3.8TTDD144 pKa = 3.03LRR146 pKa = 11.84EE147 pKa = 3.62FSMRR151 pKa = 11.84VNPVTNVPYY160 pKa = 10.38LAYY163 pKa = 10.28ALDD166 pKa = 4.15MDD168 pKa = 5.15DD169 pKa = 5.1SADD172 pKa = 3.57EE173 pKa = 4.06KK174 pKa = 11.53VGVFKK179 pKa = 11.19LDD181 pKa = 3.09GNTLVNVGAKK191 pKa = 8.58TVSEE195 pKa = 4.42GRR197 pKa = 11.84ASYY200 pKa = 10.08PRR202 pKa = 11.84IAFDD206 pKa = 4.2AEE208 pKa = 3.95GAPFISYY215 pKa = 11.16ADD217 pKa = 3.63YY218 pKa = 11.18TNNDD222 pKa = 3.68PYY224 pKa = 11.41KK225 pKa = 10.39EE226 pKa = 4.15GSTTYY231 pKa = 10.42AASVMNYY238 pKa = 9.92NGSSWSYY245 pKa = 10.66VGNKK249 pKa = 9.83GITDD253 pKa = 3.32VRR255 pKa = 11.84ITYY258 pKa = 10.78NDD260 pKa = 3.08IVLKK264 pKa = 10.93NDD266 pKa = 3.53GNPMLFCYY274 pKa = 10.61NNAAGTLARR283 pKa = 11.84RR284 pKa = 11.84EE285 pKa = 4.12LNISDD290 pKa = 3.46FTGNSWASNLTIPGRR305 pKa = 11.84ASTQYY310 pKa = 11.18AYY312 pKa = 8.73HH313 pKa = 5.63TTAKK317 pKa = 10.42NVDD320 pKa = 3.1GVIYY324 pKa = 10.51LGVYY328 pKa = 8.63NANEE332 pKa = 4.38GTFSVYY338 pKa = 10.94KK339 pKa = 9.71NTDD342 pKa = 3.36GSWTTIVEE350 pKa = 4.47SYY352 pKa = 10.42LQDD355 pKa = 3.34GATTGNLRR363 pKa = 11.84DD364 pKa = 3.51FDD366 pKa = 4.1MDD368 pKa = 3.0VDD370 pKa = 3.79RR371 pKa = 11.84DD372 pKa = 3.99GNVFVCVADD381 pKa = 4.45DD382 pKa = 3.47EE383 pKa = 5.12AGADD387 pKa = 3.15IYY389 pKa = 10.89RR390 pKa = 11.84PKK392 pKa = 9.91VLKK395 pKa = 10.7YY396 pKa = 10.57DD397 pKa = 3.54ATAEE401 pKa = 4.18TWSKK405 pKa = 11.0VGTPIAIDD413 pKa = 4.07FSTTRR418 pKa = 11.84EE419 pKa = 3.64FSLAISPVGTPFLMYY434 pKa = 9.9RR435 pKa = 11.84DD436 pKa = 4.31DD437 pKa = 4.49SMFPVVVSFDD447 pKa = 4.36EE448 pKa = 4.57DD449 pKa = 3.89TQDD452 pKa = 2.75WTTPAQLNAVEE463 pKa = 4.81AEE465 pKa = 4.02NLYY468 pKa = 10.07MDD470 pKa = 4.65FAPNGVAYY478 pKa = 10.03AAFTTSATGEE488 pKa = 4.29TMLFKK493 pKa = 10.91YY494 pKa = 9.83DD495 pKa = 3.46IPAEE499 pKa = 4.05
Molecular weight: 54.56 kDa
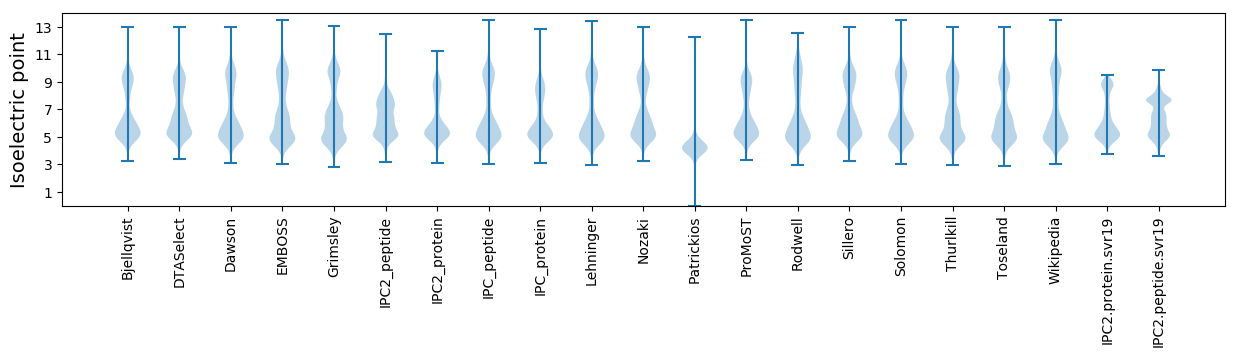
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A399SQ95|A0A399SQ95_9BACT CoA transferase (Fragment) OS=Maribellus luteus OX=2305463 GN=D1614_24595 PE=4 SV=1
AA1 pKa = 6.75QGKK4 pKa = 9.28RR5 pKa = 11.84RR6 pKa = 11.84LHH8 pKa = 6.2RR9 pKa = 11.84GGTGLSARR17 pKa = 11.84RR18 pKa = 11.84GARR21 pKa = 11.84PRR23 pKa = 11.84LGGTAPPPRR32 pKa = 11.84PPAKK36 pKa = 10.46ASGKK40 pKa = 8.58PGRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84PQLSAGLAVATGSRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84VRR63 pKa = 11.84RR64 pKa = 11.84AAPIGGPLRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.17RR76 pKa = 11.84PAPARR81 pKa = 11.84RR82 pKa = 11.84GRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84GRR88 pKa = 11.84DD89 pKa = 2.97RR90 pKa = 11.84NGRR93 pKa = 11.84RR94 pKa = 11.84SRR96 pKa = 11.84SGRR99 pKa = 11.84RR100 pKa = 11.84GPAHH104 pKa = 7.46PGRR107 pKa = 11.84HH108 pKa = 5.39PAAGRR113 pKa = 11.84GAARR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84SAGARR125 pKa = 11.84RR126 pKa = 11.84APTARR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84PLRR139 pKa = 11.84NRR141 pKa = 11.84TPGRR145 pKa = 11.84RR146 pKa = 11.84ARR148 pKa = 11.84SPRR151 pKa = 11.84RR152 pKa = 11.84SARR155 pKa = 11.84PGAGLRR161 pKa = 11.84RR162 pKa = 11.84LGPDD166 pKa = 2.43RR167 pKa = 11.84LFPEE171 pKa = 5.05RR172 pKa = 11.84PPP174 pKa = 3.72
AA1 pKa = 6.75QGKK4 pKa = 9.28RR5 pKa = 11.84RR6 pKa = 11.84LHH8 pKa = 6.2RR9 pKa = 11.84GGTGLSARR17 pKa = 11.84RR18 pKa = 11.84GARR21 pKa = 11.84PRR23 pKa = 11.84LGGTAPPPRR32 pKa = 11.84PPAKK36 pKa = 10.46ASGKK40 pKa = 8.58PGRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84PQLSAGLAVATGSRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84VRR63 pKa = 11.84RR64 pKa = 11.84AAPIGGPLRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.17RR76 pKa = 11.84PAPARR81 pKa = 11.84RR82 pKa = 11.84GRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84GRR88 pKa = 11.84DD89 pKa = 2.97RR90 pKa = 11.84NGRR93 pKa = 11.84RR94 pKa = 11.84SRR96 pKa = 11.84SGRR99 pKa = 11.84RR100 pKa = 11.84GPAHH104 pKa = 7.46PGRR107 pKa = 11.84HH108 pKa = 5.39PAAGRR113 pKa = 11.84GAARR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84SAGARR125 pKa = 11.84RR126 pKa = 11.84APTARR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84PLRR139 pKa = 11.84NRR141 pKa = 11.84TPGRR145 pKa = 11.84RR146 pKa = 11.84ARR148 pKa = 11.84SPRR151 pKa = 11.84RR152 pKa = 11.84SARR155 pKa = 11.84PGAGLRR161 pKa = 11.84RR162 pKa = 11.84LGPDD166 pKa = 2.43RR167 pKa = 11.84LFPEE171 pKa = 5.05RR172 pKa = 11.84PPP174 pKa = 3.72
Molecular weight: 19.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1801786 |
18 |
3326 |
360.4 |
40.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.962 ± 0.035 | 0.86 ± 0.012 |
5.528 ± 0.03 | 6.782 ± 0.032 |
5.052 ± 0.025 | 6.957 ± 0.031 |
1.829 ± 0.015 | 6.864 ± 0.029 |
6.693 ± 0.036 | 9.215 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.39 ± 0.014 | 5.431 ± 0.029 |
3.874 ± 0.019 | 3.508 ± 0.02 |
4.151 ± 0.023 | 6.36 ± 0.025 |
5.412 ± 0.032 | 6.641 ± 0.027 |
1.384 ± 0.014 | 4.108 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
