
Actinophytocola oryzae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Actinophytocola
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
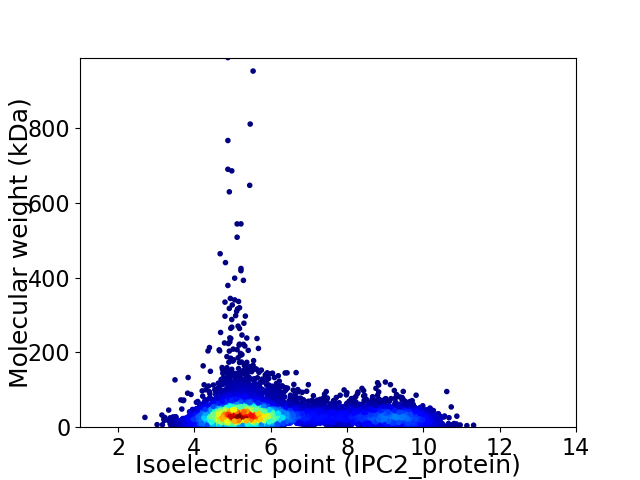
Virtual 2D-PAGE plot for 9429 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R7VQS2|A0A4R7VQS2_9PSEU Threonine/homoserine/homoserine lactone efflux protein OS=Actinophytocola oryzae OX=502181 GN=CLV71_105111 PE=4 SV=1
MM1 pKa = 7.36SRR3 pKa = 11.84RR4 pKa = 11.84LIAALGAAVVVLGLVSMPASAGAAPPPQAATAAAAQTLTWTAGDD48 pKa = 3.62SQAAYY53 pKa = 9.21TSAPTTAVAGEE64 pKa = 4.11TTIVFEE70 pKa = 4.29NSEE73 pKa = 4.18ATGNTTGMSHH83 pKa = 7.0TLTFDD88 pKa = 3.08TSTPGYY94 pKa = 10.58NNDD97 pKa = 3.13VNVNILANPFDD108 pKa = 4.98ADD110 pKa = 3.72NGHH113 pKa = 6.71HH114 pKa = 6.17EE115 pKa = 4.62VTVTLTPGKK124 pKa = 10.12YY125 pKa = 8.99RR126 pKa = 11.84YY127 pKa = 9.22HH128 pKa = 6.95CVIPGHH134 pKa = 5.23STMTGEE140 pKa = 4.25FTVTGGGGGDD150 pKa = 3.97DD151 pKa = 3.56TTAPTVTATVAGDD164 pKa = 3.39QNGDD168 pKa = 2.99GDD170 pKa = 4.74YY171 pKa = 11.08VGSATVTLDD180 pKa = 3.11ATDD183 pKa = 3.58TGGSGVDD190 pKa = 3.28SVEE193 pKa = 3.95YY194 pKa = 10.58QLDD197 pKa = 3.57GGTWTTYY204 pKa = 6.41TAPVAVSAVGDD215 pKa = 3.64HH216 pKa = 5.34TVGYY220 pKa = 9.81RR221 pKa = 11.84ATDD224 pKa = 3.37VAGNTSTEE232 pKa = 3.92GATAFTVVAGGGGDD246 pKa = 3.47TTAPTVSGSVAGAQDD261 pKa = 3.46ADD263 pKa = 3.64GNYY266 pKa = 9.87VGSATVTATAADD278 pKa = 4.08EE279 pKa = 4.74GSGVASVEE287 pKa = 3.94YY288 pKa = 10.35QINDD292 pKa = 4.11LGWNPYY298 pKa = 6.53TAPVQVTNPGQYY310 pKa = 10.1VVLLRR315 pKa = 11.84ATDD318 pKa = 3.29AAGNVGSGQVSFEE331 pKa = 4.06VVAAPSDD338 pKa = 3.8TTPPTVNAEE347 pKa = 4.14VTGDD351 pKa = 3.57QNDD354 pKa = 3.77DD355 pKa = 3.11GDD357 pKa = 4.51YY358 pKa = 11.28VDD360 pKa = 4.13VATVTLSATDD370 pKa = 3.23TGSGVKK376 pKa = 10.01SVEE379 pKa = 3.89YY380 pKa = 10.81KK381 pKa = 10.67LDD383 pKa = 4.26DD384 pKa = 4.67GAWSAYY390 pKa = 7.8TAAIPVNAPGDD401 pKa = 3.63HH402 pKa = 5.22MVGYY406 pKa = 10.2RR407 pKa = 11.84ATDD410 pKa = 3.44NANNTSDD417 pKa = 3.13EE418 pKa = 4.23GMVMFAVVQEE428 pKa = 4.67DD429 pKa = 3.28ADD431 pKa = 3.97APTVTGQVVGQQDD444 pKa = 3.36ANGAYY449 pKa = 9.79VGSAMVTLTATDD461 pKa = 3.5AGSGVASVEE470 pKa = 4.06YY471 pKa = 10.69KK472 pKa = 10.68LDD474 pKa = 3.73GGAWTAYY481 pKa = 7.22TAAVQVSTPGAHH493 pKa = 4.36TVAYY497 pKa = 9.59RR498 pKa = 11.84ATDD501 pKa = 3.22EE502 pKa = 4.65AGNVSAEE509 pKa = 4.06GTLTFTVVAGGADD522 pKa = 3.38QTPPSVSALVSGTQNSSWEE541 pKa = 4.13FLDD544 pKa = 4.25GATITVTALDD554 pKa = 3.75TGSGVGSVEE563 pKa = 3.89YY564 pKa = 10.79KK565 pKa = 10.61LDD567 pKa = 3.74GGAWTAYY574 pKa = 6.1TAPVTVDD581 pKa = 2.83SAGDD585 pKa = 3.34HH586 pKa = 5.52TFSYY590 pKa = 10.61RR591 pKa = 11.84ATDD594 pKa = 3.37KK595 pKa = 11.39AGNTSAEE602 pKa = 4.13LSGSFTVVEE611 pKa = 4.74DD612 pKa = 4.48SPGQGPDD619 pKa = 3.13VCPSSDD625 pKa = 3.35VRR627 pKa = 11.84DD628 pKa = 3.6TVYY631 pKa = 11.12VGDD634 pKa = 4.32VDD636 pKa = 4.34SQVANVDD643 pKa = 3.52TGNGCTINDD652 pKa = 4.6VIDD655 pKa = 4.92EE656 pKa = 4.31DD657 pKa = 5.74AGWQTHH663 pKa = 5.67NDD665 pKa = 3.04FVRR668 pKa = 11.84YY669 pKa = 8.77VKK671 pKa = 10.61SVTKK675 pKa = 10.3EE676 pKa = 3.61LVQNGVIDD684 pKa = 3.51NDD686 pKa = 3.06ARR688 pKa = 11.84NRR690 pKa = 11.84IVAASIDD697 pKa = 3.91STVGGEE703 pKa = 4.16AAAKK707 pKa = 9.96AA708 pKa = 3.74
MM1 pKa = 7.36SRR3 pKa = 11.84RR4 pKa = 11.84LIAALGAAVVVLGLVSMPASAGAAPPPQAATAAAAQTLTWTAGDD48 pKa = 3.62SQAAYY53 pKa = 9.21TSAPTTAVAGEE64 pKa = 4.11TTIVFEE70 pKa = 4.29NSEE73 pKa = 4.18ATGNTTGMSHH83 pKa = 7.0TLTFDD88 pKa = 3.08TSTPGYY94 pKa = 10.58NNDD97 pKa = 3.13VNVNILANPFDD108 pKa = 4.98ADD110 pKa = 3.72NGHH113 pKa = 6.71HH114 pKa = 6.17EE115 pKa = 4.62VTVTLTPGKK124 pKa = 10.12YY125 pKa = 8.99RR126 pKa = 11.84YY127 pKa = 9.22HH128 pKa = 6.95CVIPGHH134 pKa = 5.23STMTGEE140 pKa = 4.25FTVTGGGGGDD150 pKa = 3.97DD151 pKa = 3.56TTAPTVTATVAGDD164 pKa = 3.39QNGDD168 pKa = 2.99GDD170 pKa = 4.74YY171 pKa = 11.08VGSATVTLDD180 pKa = 3.11ATDD183 pKa = 3.58TGGSGVDD190 pKa = 3.28SVEE193 pKa = 3.95YY194 pKa = 10.58QLDD197 pKa = 3.57GGTWTTYY204 pKa = 6.41TAPVAVSAVGDD215 pKa = 3.64HH216 pKa = 5.34TVGYY220 pKa = 9.81RR221 pKa = 11.84ATDD224 pKa = 3.37VAGNTSTEE232 pKa = 3.92GATAFTVVAGGGGDD246 pKa = 3.47TTAPTVSGSVAGAQDD261 pKa = 3.46ADD263 pKa = 3.64GNYY266 pKa = 9.87VGSATVTATAADD278 pKa = 4.08EE279 pKa = 4.74GSGVASVEE287 pKa = 3.94YY288 pKa = 10.35QINDD292 pKa = 4.11LGWNPYY298 pKa = 6.53TAPVQVTNPGQYY310 pKa = 10.1VVLLRR315 pKa = 11.84ATDD318 pKa = 3.29AAGNVGSGQVSFEE331 pKa = 4.06VVAAPSDD338 pKa = 3.8TTPPTVNAEE347 pKa = 4.14VTGDD351 pKa = 3.57QNDD354 pKa = 3.77DD355 pKa = 3.11GDD357 pKa = 4.51YY358 pKa = 11.28VDD360 pKa = 4.13VATVTLSATDD370 pKa = 3.23TGSGVKK376 pKa = 10.01SVEE379 pKa = 3.89YY380 pKa = 10.81KK381 pKa = 10.67LDD383 pKa = 4.26DD384 pKa = 4.67GAWSAYY390 pKa = 7.8TAAIPVNAPGDD401 pKa = 3.63HH402 pKa = 5.22MVGYY406 pKa = 10.2RR407 pKa = 11.84ATDD410 pKa = 3.44NANNTSDD417 pKa = 3.13EE418 pKa = 4.23GMVMFAVVQEE428 pKa = 4.67DD429 pKa = 3.28ADD431 pKa = 3.97APTVTGQVVGQQDD444 pKa = 3.36ANGAYY449 pKa = 9.79VGSAMVTLTATDD461 pKa = 3.5AGSGVASVEE470 pKa = 4.06YY471 pKa = 10.69KK472 pKa = 10.68LDD474 pKa = 3.73GGAWTAYY481 pKa = 7.22TAAVQVSTPGAHH493 pKa = 4.36TVAYY497 pKa = 9.59RR498 pKa = 11.84ATDD501 pKa = 3.22EE502 pKa = 4.65AGNVSAEE509 pKa = 4.06GTLTFTVVAGGADD522 pKa = 3.38QTPPSVSALVSGTQNSSWEE541 pKa = 4.13FLDD544 pKa = 4.25GATITVTALDD554 pKa = 3.75TGSGVGSVEE563 pKa = 3.89YY564 pKa = 10.79KK565 pKa = 10.61LDD567 pKa = 3.74GGAWTAYY574 pKa = 6.1TAPVTVDD581 pKa = 2.83SAGDD585 pKa = 3.34HH586 pKa = 5.52TFSYY590 pKa = 10.61RR591 pKa = 11.84ATDD594 pKa = 3.37KK595 pKa = 11.39AGNTSAEE602 pKa = 4.13LSGSFTVVEE611 pKa = 4.74DD612 pKa = 4.48SPGQGPDD619 pKa = 3.13VCPSSDD625 pKa = 3.35VRR627 pKa = 11.84DD628 pKa = 3.6TVYY631 pKa = 11.12VGDD634 pKa = 4.32VDD636 pKa = 4.34SQVANVDD643 pKa = 3.52TGNGCTINDD652 pKa = 4.6VIDD655 pKa = 4.92EE656 pKa = 4.31DD657 pKa = 5.74AGWQTHH663 pKa = 5.67NDD665 pKa = 3.04FVRR668 pKa = 11.84YY669 pKa = 8.77VKK671 pKa = 10.61SVTKK675 pKa = 10.3EE676 pKa = 3.61LVQNGVIDD684 pKa = 3.51NDD686 pKa = 3.06ARR688 pKa = 11.84NRR690 pKa = 11.84IVAASIDD697 pKa = 3.91STVGGEE703 pKa = 4.16AAAKK707 pKa = 9.96AA708 pKa = 3.74
Molecular weight: 71.12 kDa
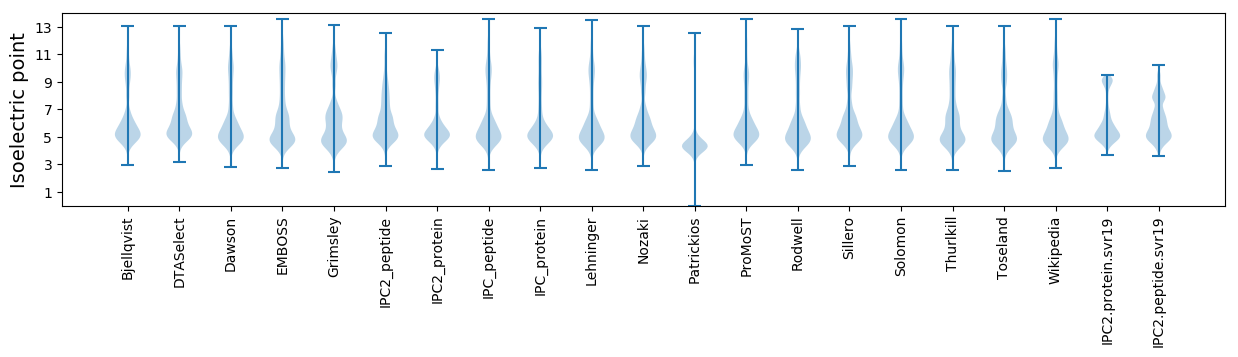
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R7VMJ9|A0A4R7VMJ9_9PSEU Sortase (Surface protein transpeptidase) OS=Actinophytocola oryzae OX=502181 GN=CLV71_106184 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILGARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.18GRR40 pKa = 11.84KK41 pKa = 8.86ALSAA45 pKa = 3.99
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILGARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.18GRR40 pKa = 11.84KK41 pKa = 8.86ALSAA45 pKa = 3.99
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3264173 |
29 |
9601 |
346.2 |
37.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.999 ± 0.043 | 0.779 ± 0.007 |
6.38 ± 0.024 | 5.325 ± 0.022 |
2.868 ± 0.014 | 9.179 ± 0.027 |
2.258 ± 0.011 | 3.125 ± 0.019 |
1.738 ± 0.02 | 10.355 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.76 ± 0.012 | 2.013 ± 0.02 |
5.899 ± 0.023 | 2.649 ± 0.018 |
7.983 ± 0.03 | 5.12 ± 0.018 |
6.514 ± 0.026 | 9.466 ± 0.032 |
1.562 ± 0.011 | 2.03 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
