
Litomosoides sigmodontis (Filarial nematode worm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Spirurina; Spiruromorpha; Filarioidea; Onchocercidae; Litomosoides
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
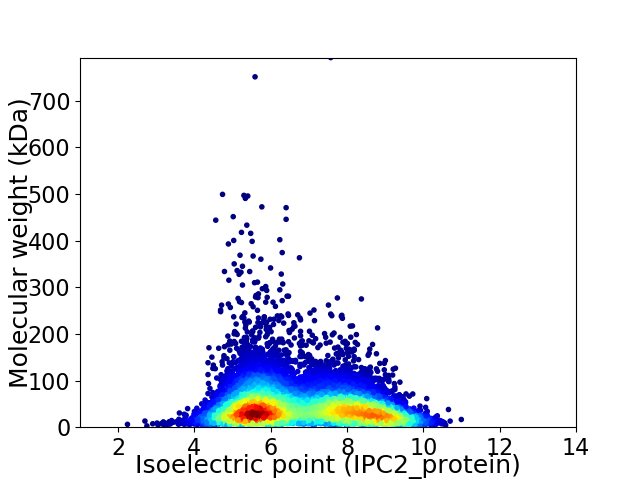
Virtual 2D-PAGE plot for 9894 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
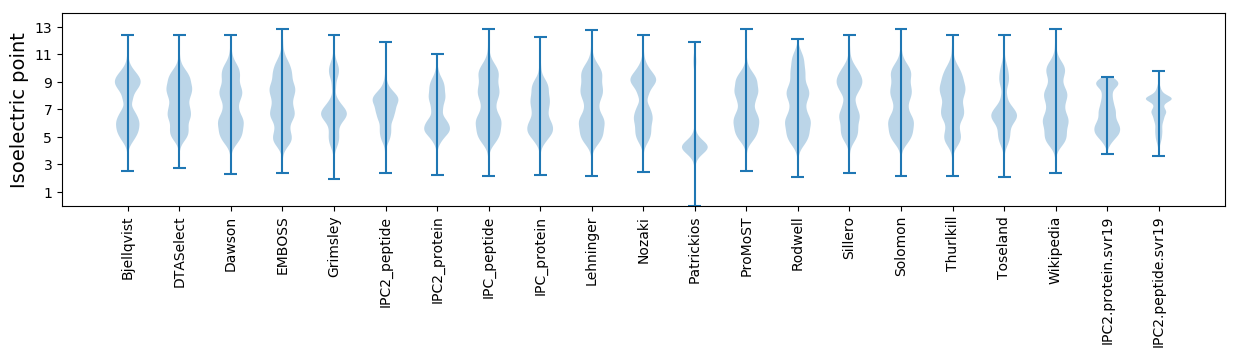
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P6T6M8|A0A3P6T6M8_LITSI PIH1 domain-containing protein OS=Litomosoides sigmodontis OX=42156 GN=NLS_LOCUS5043 PE=3 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84LIKK5 pKa = 9.48TRR7 pKa = 11.84HH8 pKa = 4.99FLRR11 pKa = 11.84VFSFYY16 pKa = 10.54LTGIHH21 pKa = 7.19SDD23 pKa = 4.06DD24 pKa = 4.91DD25 pKa = 5.29DD26 pKa = 5.79NDD28 pKa = 4.26GSDD31 pKa = 3.38VCYY34 pKa = 10.92DD35 pKa = 3.98NNNNDD40 pKa = 3.95CDD42 pKa = 4.31NDD44 pKa = 3.95NGDD47 pKa = 5.15DD48 pKa = 5.5DD49 pKa = 6.5DD50 pKa = 6.54DD51 pKa = 5.99NSDD54 pKa = 3.82NDD56 pKa = 4.0GGTDD60 pKa = 3.56DD61 pKa = 6.24DD62 pKa = 4.81EE63 pKa = 4.76WNCRR67 pKa = 11.84CNNVII72 pKa = 4.65
MM1 pKa = 7.61RR2 pKa = 11.84LIKK5 pKa = 9.48TRR7 pKa = 11.84HH8 pKa = 4.99FLRR11 pKa = 11.84VFSFYY16 pKa = 10.54LTGIHH21 pKa = 7.19SDD23 pKa = 4.06DD24 pKa = 4.91DD25 pKa = 5.29DD26 pKa = 5.79NDD28 pKa = 4.26GSDD31 pKa = 3.38VCYY34 pKa = 10.92DD35 pKa = 3.98NNNNDD40 pKa = 3.95CDD42 pKa = 4.31NDD44 pKa = 3.95NGDD47 pKa = 5.15DD48 pKa = 5.5DD49 pKa = 6.5DD50 pKa = 6.54DD51 pKa = 5.99NSDD54 pKa = 3.82NDD56 pKa = 4.0GGTDD60 pKa = 3.56DD61 pKa = 6.24DD62 pKa = 4.81EE63 pKa = 4.76WNCRR67 pKa = 11.84CNNVII72 pKa = 4.65
Molecular weight: 8.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P6SXX4|A0A3P6SXX4_LITSI Uncharacterized protein OS=Litomosoides sigmodontis OX=42156 GN=NLS_LOCUS4787 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84MMFVTPAFLSVYY14 pKa = 10.17LIAFSSLTITALFALDD30 pKa = 3.88EE31 pKa = 4.39PQWWKK36 pKa = 10.99VYY38 pKa = 9.17WIHH41 pKa = 7.57SSLLEE46 pKa = 4.31VKK48 pKa = 10.57GLLDD52 pKa = 3.56KK53 pKa = 10.69PLYY56 pKa = 10.38RR57 pKa = 11.84ILQKK61 pKa = 10.98DD62 pKa = 3.28PALINVLKK70 pKa = 10.53EE71 pKa = 3.85ALQDD75 pKa = 3.69TLVEE79 pKa = 4.29CSRR82 pKa = 11.84QSVIHH87 pKa = 6.05HH88 pKa = 6.52WKK90 pKa = 10.32CNVDD94 pKa = 3.43GAPRR98 pKa = 11.84KK99 pKa = 10.02ALFSNVASFASKK111 pKa = 10.14EE112 pKa = 3.87FAYY115 pKa = 10.67FLALSSAAAVRR126 pKa = 11.84AIAHH130 pKa = 5.63ACARR134 pKa = 11.84GRR136 pKa = 11.84LRR138 pKa = 11.84SCSCDD143 pKa = 3.23PSKK146 pKa = 10.62IGPVHH151 pKa = 7.07ADD153 pKa = 3.98QRR155 pKa = 11.84DD156 pKa = 3.17TWARR160 pKa = 11.84CSVDD164 pKa = 3.21RR165 pKa = 11.84GSDD168 pKa = 3.17NLQYY172 pKa = 11.0AVKK175 pKa = 10.23LSKK178 pKa = 10.67RR179 pKa = 11.84LIDD182 pKa = 3.48RR183 pKa = 11.84QFACSYY189 pKa = 9.4SDD191 pKa = 3.49RR192 pKa = 11.84VAQIYY197 pKa = 8.61LHH199 pKa = 6.55NIAVGRR205 pKa = 11.84SRR207 pKa = 11.84VALHH211 pKa = 6.2IKK213 pKa = 9.42CYY215 pKa = 9.86CVSAFGSCRR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84HH227 pKa = 5.98CEE229 pKa = 3.52AKK231 pKa = 10.17VVPFEE236 pKa = 4.16MTGDD240 pKa = 3.55AVMKK244 pKa = 10.41SLLRR248 pKa = 11.84SRR250 pKa = 11.84RR251 pKa = 11.84IRR253 pKa = 11.84RR254 pKa = 11.84SNSLPTTRR262 pKa = 11.84VTCLRR267 pKa = 11.84PRR269 pKa = 11.84NEE271 pKa = 3.77RR272 pKa = 11.84NMRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84VVPLWFIDD285 pKa = 3.51TRR287 pKa = 11.84SWTMRR292 pKa = 11.84SGKK295 pKa = 7.96SRR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84LKK301 pKa = 10.82NN302 pKa = 3.05
MM1 pKa = 8.06RR2 pKa = 11.84MMFVTPAFLSVYY14 pKa = 10.17LIAFSSLTITALFALDD30 pKa = 3.88EE31 pKa = 4.39PQWWKK36 pKa = 10.99VYY38 pKa = 9.17WIHH41 pKa = 7.57SSLLEE46 pKa = 4.31VKK48 pKa = 10.57GLLDD52 pKa = 3.56KK53 pKa = 10.69PLYY56 pKa = 10.38RR57 pKa = 11.84ILQKK61 pKa = 10.98DD62 pKa = 3.28PALINVLKK70 pKa = 10.53EE71 pKa = 3.85ALQDD75 pKa = 3.69TLVEE79 pKa = 4.29CSRR82 pKa = 11.84QSVIHH87 pKa = 6.05HH88 pKa = 6.52WKK90 pKa = 10.32CNVDD94 pKa = 3.43GAPRR98 pKa = 11.84KK99 pKa = 10.02ALFSNVASFASKK111 pKa = 10.14EE112 pKa = 3.87FAYY115 pKa = 10.67FLALSSAAAVRR126 pKa = 11.84AIAHH130 pKa = 5.63ACARR134 pKa = 11.84GRR136 pKa = 11.84LRR138 pKa = 11.84SCSCDD143 pKa = 3.23PSKK146 pKa = 10.62IGPVHH151 pKa = 7.07ADD153 pKa = 3.98QRR155 pKa = 11.84DD156 pKa = 3.17TWARR160 pKa = 11.84CSVDD164 pKa = 3.21RR165 pKa = 11.84GSDD168 pKa = 3.17NLQYY172 pKa = 11.0AVKK175 pKa = 10.23LSKK178 pKa = 10.67RR179 pKa = 11.84LIDD182 pKa = 3.48RR183 pKa = 11.84QFACSYY189 pKa = 9.4SDD191 pKa = 3.49RR192 pKa = 11.84VAQIYY197 pKa = 8.61LHH199 pKa = 6.55NIAVGRR205 pKa = 11.84SRR207 pKa = 11.84VALHH211 pKa = 6.2IKK213 pKa = 9.42CYY215 pKa = 9.86CVSAFGSCRR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84HH227 pKa = 5.98CEE229 pKa = 3.52AKK231 pKa = 10.17VVPFEE236 pKa = 4.16MTGDD240 pKa = 3.55AVMKK244 pKa = 10.41SLLRR248 pKa = 11.84SRR250 pKa = 11.84RR251 pKa = 11.84IRR253 pKa = 11.84RR254 pKa = 11.84SNSLPTTRR262 pKa = 11.84VTCLRR267 pKa = 11.84PRR269 pKa = 11.84NEE271 pKa = 3.77RR272 pKa = 11.84NMRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84VVPLWFIDD285 pKa = 3.51TRR287 pKa = 11.84SWTMRR292 pKa = 11.84SGKK295 pKa = 7.96SRR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84LKK301 pKa = 10.82NN302 pKa = 3.05
Molecular weight: 34.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4320305 |
10 |
7030 |
436.7 |
49.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.528 ± 0.02 | 2.236 ± 0.018 |
5.466 ± 0.016 | 6.671 ± 0.026 |
4.096 ± 0.016 | 5.258 ± 0.025 |
2.414 ± 0.009 | 6.082 ± 0.016 |
6.021 ± 0.021 | 9.26 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.012 | 4.863 ± 0.014 |
4.293 ± 0.023 | 4.094 ± 0.019 |
5.934 ± 0.02 | 8.357 ± 0.027 |
5.508 ± 0.02 | 6.196 ± 0.016 |
1.1 ± 0.008 | 3.099 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
