
Fusarium oxysporum f. sp. cubense (strain race 4) (Panama disease fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Nectriaceae; Fusarium; Fusarium oxysporum species complex
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
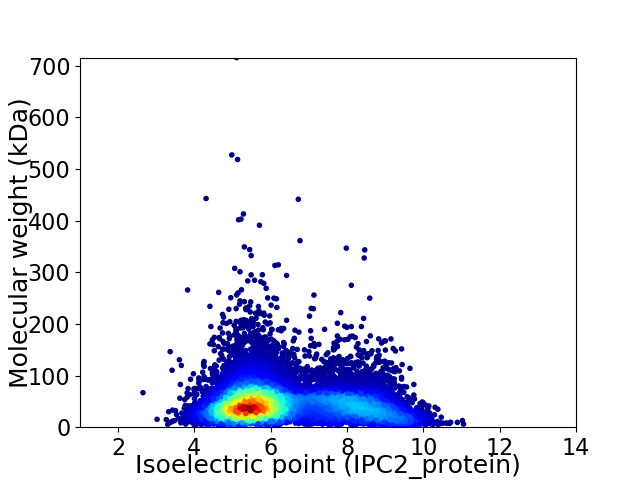
Virtual 2D-PAGE plot for 14157 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|N1R907|N1R907_FUSC4 Histidine biosynthesis trifunctional protein OS=Fusarium oxysporum f. sp. cubense (strain race 4) OX=2502994 GN=FOC4_g10014644 PE=3 SV=1
MM1 pKa = 7.78KK2 pKa = 10.45SQILLTTAMGASGVLSQAGHH22 pKa = 6.32GAMSLDD28 pKa = 3.91TWCVTYY34 pKa = 10.81LSTYY38 pKa = 9.42LVPVANPDD46 pKa = 3.06QATDD50 pKa = 3.56VSSSQPPFTGNSSVPTDD67 pKa = 3.51CVTEE71 pKa = 4.3TEE73 pKa = 4.4SQSALTSDD81 pKa = 3.38TSVFTSVAVSSSVEE95 pKa = 4.1TEE97 pKa = 3.74LTEE100 pKa = 4.22AQSGSEE106 pKa = 4.05TVSFDD111 pKa = 3.32STAGTLADD119 pKa = 4.2QITSASSEE127 pKa = 4.32STISTDD133 pKa = 3.73ALTTSTSIIEE143 pKa = 4.01PPGRR147 pKa = 11.84SVIFLISAPNTRR159 pKa = 11.84KK160 pKa = 9.85RR161 pKa = 11.84QNTDD165 pKa = 2.14KK166 pKa = 11.42DD167 pKa = 3.86GVLFFYY173 pKa = 10.82DD174 pKa = 3.44GEE176 pKa = 4.46DD177 pKa = 3.51YY178 pKa = 11.15KK179 pKa = 11.12EE180 pKa = 4.24LVMGPTPPAGAVTRR194 pKa = 11.84LFTTAGRR201 pKa = 11.84SLQVDD206 pKa = 4.55LPGGEE211 pKa = 4.95AGFCQTSDD219 pKa = 2.56GRR221 pKa = 11.84VYY223 pKa = 10.12VTFTRR228 pKa = 11.84GPAGCEE234 pKa = 3.52AVSLDD239 pKa = 3.93VYY241 pKa = 10.62DD242 pKa = 5.61EE243 pKa = 4.32RR244 pKa = 11.84QCQNGRR250 pKa = 11.84LVGIDD255 pKa = 3.34TTTSAMKK262 pKa = 9.12TATSEE267 pKa = 4.38VISSGNATSAEE278 pKa = 4.06GSATTEE284 pKa = 4.07TTEE287 pKa = 4.21TTSEE291 pKa = 4.31SIATSSNVAQTQSVGPVSQTTHH313 pKa = 6.7SEE315 pKa = 3.72ASTAASSRR323 pKa = 11.84AVEE326 pKa = 4.22SSTSVLSTTTTTTVVSDD343 pKa = 5.12ASTQSQASSSVLGEE357 pKa = 4.08STTDD361 pKa = 3.18EE362 pKa = 4.47VFPTTAPSLVSTSAQAEE379 pKa = 4.2EE380 pKa = 4.34STSIKK385 pKa = 10.11PSSQASSEE393 pKa = 4.26TTEE396 pKa = 4.02SGTTSSEE403 pKa = 3.74EE404 pKa = 4.11STSDD408 pKa = 3.23VALEE412 pKa = 4.21TTTEE416 pKa = 3.83ARR418 pKa = 11.84TSTRR422 pKa = 11.84EE423 pKa = 3.48TTAADD428 pKa = 3.49EE429 pKa = 4.35TSTEE433 pKa = 4.1NEE435 pKa = 4.31TTTEE439 pKa = 3.84AATEE443 pKa = 4.33SEE445 pKa = 4.48TTTTEE450 pKa = 3.56SSAPSEE456 pKa = 5.09DD457 pKa = 3.31ISTTDD462 pKa = 2.98VTAVDD467 pKa = 3.81SSTAATTTTAALACTDD483 pKa = 3.98LSDD486 pKa = 4.9PYY488 pKa = 10.16MDD490 pKa = 3.89STGTTYY496 pKa = 11.64ALLCNTDD503 pKa = 3.15VNGYY507 pKa = 7.22TSINGFTAASFVACIEE523 pKa = 4.06ACSVYY528 pKa = 10.52VGCAGIEE535 pKa = 4.07FLKK538 pKa = 10.06STGYY542 pKa = 8.42CTLFNSSTGSSPTTDD557 pKa = 2.96YY558 pKa = 11.42DD559 pKa = 3.35IALGALRR566 pKa = 11.84EE567 pKa = 4.33GG568 pKa = 3.89
MM1 pKa = 7.78KK2 pKa = 10.45SQILLTTAMGASGVLSQAGHH22 pKa = 6.32GAMSLDD28 pKa = 3.91TWCVTYY34 pKa = 10.81LSTYY38 pKa = 9.42LVPVANPDD46 pKa = 3.06QATDD50 pKa = 3.56VSSSQPPFTGNSSVPTDD67 pKa = 3.51CVTEE71 pKa = 4.3TEE73 pKa = 4.4SQSALTSDD81 pKa = 3.38TSVFTSVAVSSSVEE95 pKa = 4.1TEE97 pKa = 3.74LTEE100 pKa = 4.22AQSGSEE106 pKa = 4.05TVSFDD111 pKa = 3.32STAGTLADD119 pKa = 4.2QITSASSEE127 pKa = 4.32STISTDD133 pKa = 3.73ALTTSTSIIEE143 pKa = 4.01PPGRR147 pKa = 11.84SVIFLISAPNTRR159 pKa = 11.84KK160 pKa = 9.85RR161 pKa = 11.84QNTDD165 pKa = 2.14KK166 pKa = 11.42DD167 pKa = 3.86GVLFFYY173 pKa = 10.82DD174 pKa = 3.44GEE176 pKa = 4.46DD177 pKa = 3.51YY178 pKa = 11.15KK179 pKa = 11.12EE180 pKa = 4.24LVMGPTPPAGAVTRR194 pKa = 11.84LFTTAGRR201 pKa = 11.84SLQVDD206 pKa = 4.55LPGGEE211 pKa = 4.95AGFCQTSDD219 pKa = 2.56GRR221 pKa = 11.84VYY223 pKa = 10.12VTFTRR228 pKa = 11.84GPAGCEE234 pKa = 3.52AVSLDD239 pKa = 3.93VYY241 pKa = 10.62DD242 pKa = 5.61EE243 pKa = 4.32RR244 pKa = 11.84QCQNGRR250 pKa = 11.84LVGIDD255 pKa = 3.34TTTSAMKK262 pKa = 9.12TATSEE267 pKa = 4.38VISSGNATSAEE278 pKa = 4.06GSATTEE284 pKa = 4.07TTEE287 pKa = 4.21TTSEE291 pKa = 4.31SIATSSNVAQTQSVGPVSQTTHH313 pKa = 6.7SEE315 pKa = 3.72ASTAASSRR323 pKa = 11.84AVEE326 pKa = 4.22SSTSVLSTTTTTTVVSDD343 pKa = 5.12ASTQSQASSSVLGEE357 pKa = 4.08STTDD361 pKa = 3.18EE362 pKa = 4.47VFPTTAPSLVSTSAQAEE379 pKa = 4.2EE380 pKa = 4.34STSIKK385 pKa = 10.11PSSQASSEE393 pKa = 4.26TTEE396 pKa = 4.02SGTTSSEE403 pKa = 3.74EE404 pKa = 4.11STSDD408 pKa = 3.23VALEE412 pKa = 4.21TTTEE416 pKa = 3.83ARR418 pKa = 11.84TSTRR422 pKa = 11.84EE423 pKa = 3.48TTAADD428 pKa = 3.49EE429 pKa = 4.35TSTEE433 pKa = 4.1NEE435 pKa = 4.31TTTEE439 pKa = 3.84AATEE443 pKa = 4.33SEE445 pKa = 4.48TTTTEE450 pKa = 3.56SSAPSEE456 pKa = 5.09DD457 pKa = 3.31ISTTDD462 pKa = 2.98VTAVDD467 pKa = 3.81SSTAATTTTAALACTDD483 pKa = 3.98LSDD486 pKa = 4.9PYY488 pKa = 10.16MDD490 pKa = 3.89STGTTYY496 pKa = 11.64ALLCNTDD503 pKa = 3.15VNGYY507 pKa = 7.22TSINGFTAASFVACIEE523 pKa = 4.06ACSVYY528 pKa = 10.52VGCAGIEE535 pKa = 4.07FLKK538 pKa = 10.06STGYY542 pKa = 8.42CTLFNSSTGSSPTTDD557 pKa = 2.96YY558 pKa = 11.42DD559 pKa = 3.35IALGALRR566 pKa = 11.84EE567 pKa = 4.33GG568 pKa = 3.89
Molecular weight: 58.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N1RB97|N1RB97_FUSC4 HET domain-containing protein (Fragment) OS=Fusarium oxysporum f. sp. cubense (strain race 4) OX=2502994 GN=FOC4_g10006368 PE=4 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APAHH26 pKa = 4.84SHH28 pKa = 5.33RR29 pKa = 11.84HH30 pKa = 4.03TTTTTTTTMKK40 pKa = 9.74PRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GMFGGGAGRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 6.46ATTTAPVHH64 pKa = 4.87HH65 pKa = 6.63HH66 pKa = 5.14QRR68 pKa = 11.84RR69 pKa = 11.84PSMKK73 pKa = 10.02DD74 pKa = 2.95KK75 pKa = 11.41VSGALLKK82 pKa = 11.04LKK84 pKa = 10.68GSLTRR89 pKa = 11.84RR90 pKa = 11.84PGVKK94 pKa = 9.89AAGTRR99 pKa = 11.84RR100 pKa = 11.84MRR102 pKa = 11.84GTDD105 pKa = 3.02GRR107 pKa = 11.84GARR110 pKa = 11.84HH111 pKa = 5.87HH112 pKa = 7.23RR113 pKa = 11.84YY114 pKa = 9.44
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APAHH26 pKa = 4.84SHH28 pKa = 5.33RR29 pKa = 11.84HH30 pKa = 4.03TTTTTTTTMKK40 pKa = 9.74PRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GMFGGGAGRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 6.46ATTTAPVHH64 pKa = 4.87HH65 pKa = 6.63HH66 pKa = 5.14QRR68 pKa = 11.84RR69 pKa = 11.84PSMKK73 pKa = 10.02DD74 pKa = 2.95KK75 pKa = 11.41VSGALLKK82 pKa = 11.04LKK84 pKa = 10.68GSLTRR89 pKa = 11.84RR90 pKa = 11.84PGVKK94 pKa = 9.89AAGTRR99 pKa = 11.84RR100 pKa = 11.84MRR102 pKa = 11.84GTDD105 pKa = 3.02GRR107 pKa = 11.84GARR110 pKa = 11.84HH111 pKa = 5.87HH112 pKa = 7.23RR113 pKa = 11.84YY114 pKa = 9.44
Molecular weight: 12.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6288100 |
26 |
6475 |
444.2 |
49.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.354 ± 0.019 | 1.319 ± 0.009 |
5.8 ± 0.016 | 6.151 ± 0.021 |
3.845 ± 0.013 | 6.812 ± 0.02 |
2.368 ± 0.01 | 5.179 ± 0.015 |
5.038 ± 0.018 | 8.865 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.244 ± 0.008 | 3.776 ± 0.01 |
5.78 ± 0.022 | 3.947 ± 0.014 |
5.781 ± 0.017 | 7.962 ± 0.021 |
5.962 ± 0.019 | 6.113 ± 0.013 |
1.587 ± 0.008 | 2.842 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
