
Corynebacterium cystitidis DSM 20524
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium; Corynebacterium cystitidis
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
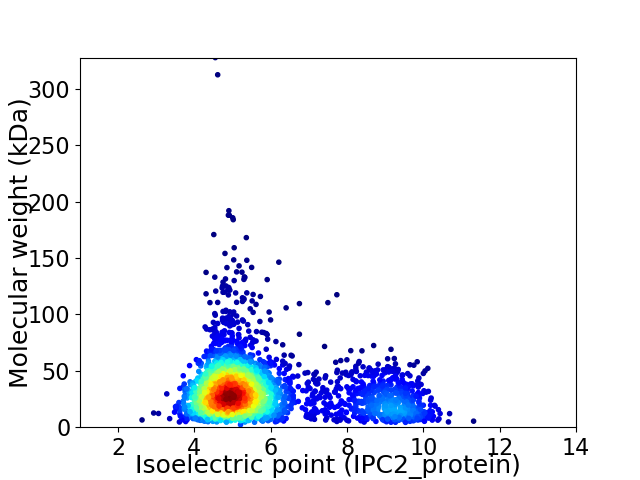
Virtual 2D-PAGE plot for 2787 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
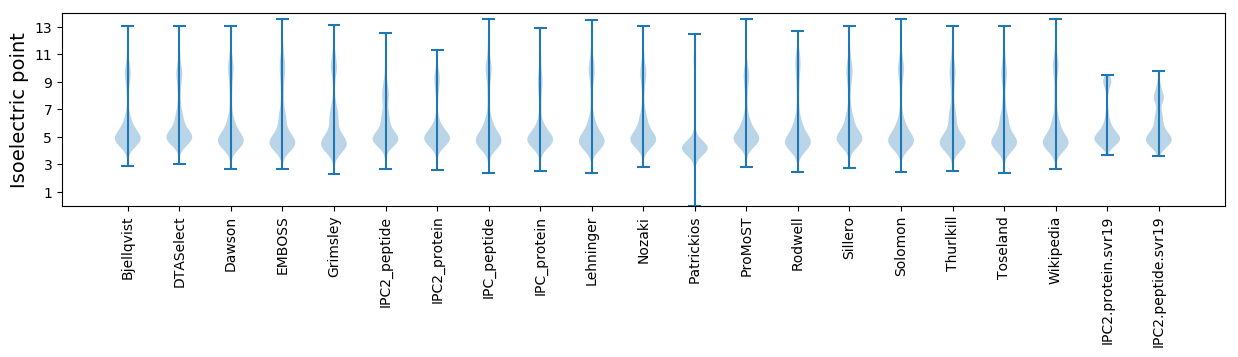
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9VG85|A0A1H9VG85_9CORY Long-chain acyl-CoA synthetase OS=Corynebacterium cystitidis DSM 20524 OX=1121357 GN=SAMN05661109_02227 PE=4 SV=1
MM1 pKa = 7.72SDD3 pKa = 3.64DD4 pKa = 3.89TSCDD8 pKa = 2.97GLQFNRR14 pKa = 11.84TPVYY18 pKa = 9.83ISLAIAIVLVVAVLFGAKK36 pKa = 8.62WYY38 pKa = 10.42FDD40 pKa = 3.55QATNTPVAMGAVDD53 pKa = 4.5APEE56 pKa = 4.72ADD58 pKa = 3.76SPQCQSLIDD67 pKa = 4.44DD68 pKa = 5.14LPDD71 pKa = 3.35EE72 pKa = 4.8LLGHH76 pKa = 6.93DD77 pKa = 3.74RR78 pKa = 11.84AEE80 pKa = 4.14LTEE83 pKa = 4.33PKK85 pKa = 9.9PAGAAAWQTTEE96 pKa = 3.85IEE98 pKa = 4.8RR99 pKa = 11.84ITLRR103 pKa = 11.84CGVAVPLQYY112 pKa = 10.83DD113 pKa = 4.39EE114 pKa = 4.53YY115 pKa = 11.37AQTEE119 pKa = 4.32QINGVEE125 pKa = 4.22WLRR128 pKa = 11.84VDD130 pKa = 4.97DD131 pKa = 4.18YY132 pKa = 11.28TPEE135 pKa = 4.24STLSTWYY142 pKa = 9.29TVDD145 pKa = 3.59RR146 pKa = 11.84YY147 pKa = 10.23PVVAVTADD155 pKa = 3.36DD156 pKa = 4.02HH157 pKa = 7.24ALGGAEE163 pKa = 3.83NPVEE167 pKa = 4.05NVPVGQLEE175 pKa = 4.02QRR177 pKa = 11.84EE178 pKa = 4.41VEE180 pKa = 4.73PYY182 pKa = 8.92PAPLSNLAAGDD193 pKa = 3.68ASVAGACATLMEE205 pKa = 4.82TLPEE209 pKa = 4.3DD210 pKa = 3.69VADD213 pKa = 5.0GYY215 pKa = 11.7SPLAANTGGAEE226 pKa = 4.38LDD228 pKa = 3.69PDD230 pKa = 4.19TAVWTAPGAEE240 pKa = 4.32SIVLRR245 pKa = 11.84CGVADD250 pKa = 3.91PEE252 pKa = 4.26NYY254 pKa = 9.58QPGEE258 pKa = 3.89QLHH261 pKa = 6.06QINDD265 pKa = 3.64VTWFEE270 pKa = 4.16DD271 pKa = 3.48TQLINGSTASTWFALGRR288 pKa = 11.84EE289 pKa = 4.41ANVAVSMPQFVGNSAIVRR307 pKa = 11.84ITEE310 pKa = 4.75AIEE313 pKa = 3.86KK314 pKa = 10.07ALPAEE319 pKa = 4.32
MM1 pKa = 7.72SDD3 pKa = 3.64DD4 pKa = 3.89TSCDD8 pKa = 2.97GLQFNRR14 pKa = 11.84TPVYY18 pKa = 9.83ISLAIAIVLVVAVLFGAKK36 pKa = 8.62WYY38 pKa = 10.42FDD40 pKa = 3.55QATNTPVAMGAVDD53 pKa = 4.5APEE56 pKa = 4.72ADD58 pKa = 3.76SPQCQSLIDD67 pKa = 4.44DD68 pKa = 5.14LPDD71 pKa = 3.35EE72 pKa = 4.8LLGHH76 pKa = 6.93DD77 pKa = 3.74RR78 pKa = 11.84AEE80 pKa = 4.14LTEE83 pKa = 4.33PKK85 pKa = 9.9PAGAAAWQTTEE96 pKa = 3.85IEE98 pKa = 4.8RR99 pKa = 11.84ITLRR103 pKa = 11.84CGVAVPLQYY112 pKa = 10.83DD113 pKa = 4.39EE114 pKa = 4.53YY115 pKa = 11.37AQTEE119 pKa = 4.32QINGVEE125 pKa = 4.22WLRR128 pKa = 11.84VDD130 pKa = 4.97DD131 pKa = 4.18YY132 pKa = 11.28TPEE135 pKa = 4.24STLSTWYY142 pKa = 9.29TVDD145 pKa = 3.59RR146 pKa = 11.84YY147 pKa = 10.23PVVAVTADD155 pKa = 3.36DD156 pKa = 4.02HH157 pKa = 7.24ALGGAEE163 pKa = 3.83NPVEE167 pKa = 4.05NVPVGQLEE175 pKa = 4.02QRR177 pKa = 11.84EE178 pKa = 4.41VEE180 pKa = 4.73PYY182 pKa = 8.92PAPLSNLAAGDD193 pKa = 3.68ASVAGACATLMEE205 pKa = 4.82TLPEE209 pKa = 4.3DD210 pKa = 3.69VADD213 pKa = 5.0GYY215 pKa = 11.7SPLAANTGGAEE226 pKa = 4.38LDD228 pKa = 3.69PDD230 pKa = 4.19TAVWTAPGAEE240 pKa = 4.32SIVLRR245 pKa = 11.84CGVADD250 pKa = 3.91PEE252 pKa = 4.26NYY254 pKa = 9.58QPGEE258 pKa = 3.89QLHH261 pKa = 6.06QINDD265 pKa = 3.64VTWFEE270 pKa = 4.16DD271 pKa = 3.48TQLINGSTASTWFALGRR288 pKa = 11.84EE289 pKa = 4.41ANVAVSMPQFVGNSAIVRR307 pKa = 11.84ITEE310 pKa = 4.75AIEE313 pKa = 3.86KK314 pKa = 10.07ALPAEE319 pKa = 4.32
Molecular weight: 34.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9UMT5|A0A1H9UMT5_9CORY Non-specific serine/threonine protein kinase OS=Corynebacterium cystitidis DSM 20524 OX=1121357 GN=SAMN05661109_01882 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84KK16 pKa = 8.53HH17 pKa = 4.82GFRR20 pKa = 11.84TRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVSARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.64KK38 pKa = 10.52GRR40 pKa = 11.84AKK42 pKa = 9.67LTAA45 pKa = 4.21
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84KK16 pKa = 8.53HH17 pKa = 4.82GFRR20 pKa = 11.84TRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVSARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.64KK38 pKa = 10.52GRR40 pKa = 11.84AKK42 pKa = 9.67LTAA45 pKa = 4.21
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
873279 |
19 |
3087 |
313.3 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.083 ± 0.055 | 0.655 ± 0.011 |
6.262 ± 0.042 | 6.262 ± 0.046 |
3.357 ± 0.031 | 8.207 ± 0.043 |
2.237 ± 0.021 | 5.162 ± 0.035 |
3.182 ± 0.036 | 9.375 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.254 ± 0.02 | 3.044 ± 0.025 |
5.008 ± 0.034 | 3.492 ± 0.024 |
6.019 ± 0.04 | 5.88 ± 0.037 |
6.367 ± 0.034 | 8.332 ± 0.041 |
1.454 ± 0.02 | 2.368 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
