
marine gamma proteobacterium HTCC2143
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Spongiibacteraceae; unclassified Spongiibacteraceae; BD1-7 clade
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
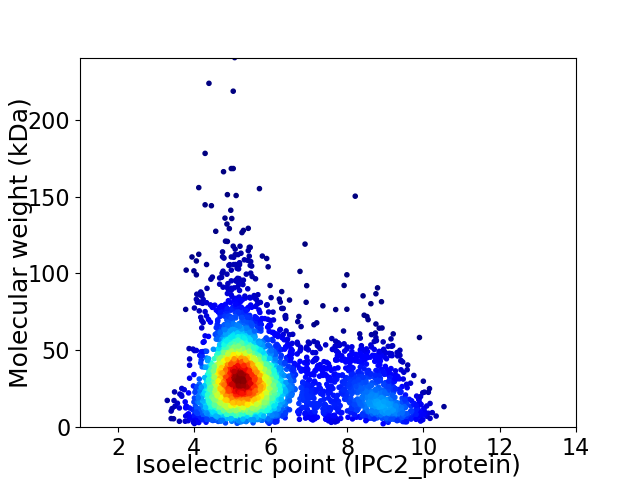
Virtual 2D-PAGE plot for 3661 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0YFZ0|A0YFZ0_9GAMM Uncharacterized protein OS=marine gamma proteobacterium HTCC2143 OX=247633 GN=GP2143_01815 PE=4 SV=1
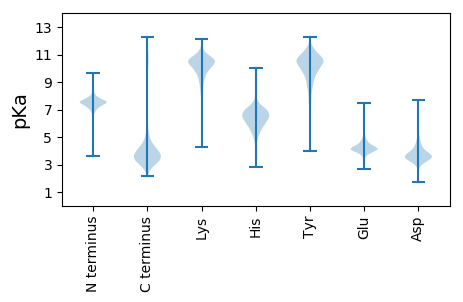
GG1 pKa = 7.44GDD3 pKa = 3.11VDD5 pKa = 4.2YY6 pKa = 10.8TSLDD10 pKa = 3.08IVRR13 pKa = 11.84TEE15 pKa = 4.14DD16 pKa = 3.2SLEE19 pKa = 4.26IEE21 pKa = 4.51TVTQEE26 pKa = 4.19FRR28 pKa = 11.84LTSTAGEE35 pKa = 4.25SFDD38 pKa = 3.37WMVGGFIFQEE48 pKa = 4.7DD49 pKa = 4.17IEE51 pKa = 4.77TDD53 pKa = 3.11GSTFFGNDD61 pKa = 2.3FRR63 pKa = 11.84AYY65 pKa = 10.57GDD67 pKa = 3.53VLFGAFPNILSTVEE81 pKa = 4.21LLSGLPAGAILSSDD95 pKa = 3.4NVASTAFEE103 pKa = 4.01QDD105 pKa = 2.66NFAYY109 pKa = 10.63SLFATGDD116 pKa = 3.29YY117 pKa = 10.95HH118 pKa = 6.8FTDD121 pKa = 4.19KK122 pKa = 10.4LTATLGVSYY131 pKa = 9.63TYY133 pKa = 10.51DD134 pKa = 3.19EE135 pKa = 4.87KK136 pKa = 11.44EE137 pKa = 4.3VVKK140 pKa = 10.79DD141 pKa = 3.22SYY143 pKa = 11.6RR144 pKa = 11.84NNDD147 pKa = 3.34LYY149 pKa = 11.5SAVDD153 pKa = 3.77LVPVLGATPLAGLIPSLQGLQFRR176 pKa = 11.84PPVIDD181 pKa = 3.51YY182 pKa = 8.85PNSVEE187 pKa = 4.61NGEE190 pKa = 4.19SDD192 pKa = 3.75DD193 pKa = 6.55DD194 pKa = 4.1KK195 pKa = 11.2VTWTARR201 pKa = 11.84VTYY204 pKa = 9.65QVNDD208 pKa = 3.6MVNVYY213 pKa = 10.57ASAATGYY220 pKa = 10.49KK221 pKa = 9.83SSSWNIGGASRR232 pKa = 11.84PFPASEE238 pKa = 4.07AALDD242 pKa = 3.86AAGLLVANQSFGTRR256 pKa = 11.84FASPEE261 pKa = 3.62EE262 pKa = 3.83AMVYY266 pKa = 10.36EE267 pKa = 5.02LGLKK271 pKa = 10.42ASFEE275 pKa = 4.03QGAFNIAIFDD285 pKa = 3.57QTIEE289 pKa = 4.75DD290 pKa = 4.06FQQNAFDD297 pKa = 4.46GDD299 pKa = 4.36SFVLTNAGEE308 pKa = 3.95RR309 pKa = 11.84STRR312 pKa = 11.84GIEE315 pKa = 3.82FDD317 pKa = 3.77ALYY320 pKa = 10.78SPTDD324 pKa = 2.75AWLFNLAGTFMDD336 pKa = 4.75PKK338 pKa = 9.77YY339 pKa = 10.89DD340 pKa = 3.59SYY342 pKa = 11.55TGASGIGGAPIDD354 pKa = 4.27LSGEE358 pKa = 4.08EE359 pKa = 3.69PAGIHH364 pKa = 6.12KK365 pKa = 10.72EE366 pKa = 4.01NVTASATYY374 pKa = 10.36RR375 pKa = 11.84FTVGDD380 pKa = 4.75GITGYY385 pKa = 10.67VRR387 pKa = 11.84TDD389 pKa = 3.28YY390 pKa = 10.61IYY392 pKa = 10.84EE393 pKa = 4.18SDD395 pKa = 5.48VRR397 pKa = 11.84AAWNVPEE404 pKa = 4.16EE405 pKa = 4.2FSRR408 pKa = 11.84EE409 pKa = 3.85VSTWNASAGLQFTNGVNVQVWGRR432 pKa = 11.84NINDD436 pKa = 3.42DD437 pKa = 3.76EE438 pKa = 5.51YY439 pKa = 11.49YY440 pKa = 10.6VGVFQGVVQSGTYY453 pKa = 9.78NAFASAPATYY463 pKa = 9.68GVNVSYY469 pKa = 10.9EE470 pKa = 3.98FF471 pKa = 4.22
GG1 pKa = 7.44GDD3 pKa = 3.11VDD5 pKa = 4.2YY6 pKa = 10.8TSLDD10 pKa = 3.08IVRR13 pKa = 11.84TEE15 pKa = 4.14DD16 pKa = 3.2SLEE19 pKa = 4.26IEE21 pKa = 4.51TVTQEE26 pKa = 4.19FRR28 pKa = 11.84LTSTAGEE35 pKa = 4.25SFDD38 pKa = 3.37WMVGGFIFQEE48 pKa = 4.7DD49 pKa = 4.17IEE51 pKa = 4.77TDD53 pKa = 3.11GSTFFGNDD61 pKa = 2.3FRR63 pKa = 11.84AYY65 pKa = 10.57GDD67 pKa = 3.53VLFGAFPNILSTVEE81 pKa = 4.21LLSGLPAGAILSSDD95 pKa = 3.4NVASTAFEE103 pKa = 4.01QDD105 pKa = 2.66NFAYY109 pKa = 10.63SLFATGDD116 pKa = 3.29YY117 pKa = 10.95HH118 pKa = 6.8FTDD121 pKa = 4.19KK122 pKa = 10.4LTATLGVSYY131 pKa = 9.63TYY133 pKa = 10.51DD134 pKa = 3.19EE135 pKa = 4.87KK136 pKa = 11.44EE137 pKa = 4.3VVKK140 pKa = 10.79DD141 pKa = 3.22SYY143 pKa = 11.6RR144 pKa = 11.84NNDD147 pKa = 3.34LYY149 pKa = 11.5SAVDD153 pKa = 3.77LVPVLGATPLAGLIPSLQGLQFRR176 pKa = 11.84PPVIDD181 pKa = 3.51YY182 pKa = 8.85PNSVEE187 pKa = 4.61NGEE190 pKa = 4.19SDD192 pKa = 3.75DD193 pKa = 6.55DD194 pKa = 4.1KK195 pKa = 11.2VTWTARR201 pKa = 11.84VTYY204 pKa = 9.65QVNDD208 pKa = 3.6MVNVYY213 pKa = 10.57ASAATGYY220 pKa = 10.49KK221 pKa = 9.83SSSWNIGGASRR232 pKa = 11.84PFPASEE238 pKa = 4.07AALDD242 pKa = 3.86AAGLLVANQSFGTRR256 pKa = 11.84FASPEE261 pKa = 3.62EE262 pKa = 3.83AMVYY266 pKa = 10.36EE267 pKa = 5.02LGLKK271 pKa = 10.42ASFEE275 pKa = 4.03QGAFNIAIFDD285 pKa = 3.57QTIEE289 pKa = 4.75DD290 pKa = 4.06FQQNAFDD297 pKa = 4.46GDD299 pKa = 4.36SFVLTNAGEE308 pKa = 3.95RR309 pKa = 11.84STRR312 pKa = 11.84GIEE315 pKa = 3.82FDD317 pKa = 3.77ALYY320 pKa = 10.78SPTDD324 pKa = 2.75AWLFNLAGTFMDD336 pKa = 4.75PKK338 pKa = 9.77YY339 pKa = 10.89DD340 pKa = 3.59SYY342 pKa = 11.55TGASGIGGAPIDD354 pKa = 4.27LSGEE358 pKa = 4.08EE359 pKa = 3.69PAGIHH364 pKa = 6.12KK365 pKa = 10.72EE366 pKa = 4.01NVTASATYY374 pKa = 10.36RR375 pKa = 11.84FTVGDD380 pKa = 4.75GITGYY385 pKa = 10.67VRR387 pKa = 11.84TDD389 pKa = 3.28YY390 pKa = 10.61IYY392 pKa = 10.84EE393 pKa = 4.18SDD395 pKa = 5.48VRR397 pKa = 11.84AAWNVPEE404 pKa = 4.16EE405 pKa = 4.2FSRR408 pKa = 11.84EE409 pKa = 3.85VSTWNASAGLQFTNGVNVQVWGRR432 pKa = 11.84NINDD436 pKa = 3.42DD437 pKa = 3.76EE438 pKa = 5.51YY439 pKa = 11.49YY440 pKa = 10.6VGVFQGVVQSGTYY453 pKa = 9.78NAFASAPATYY463 pKa = 9.68GVNVSYY469 pKa = 10.9EE470 pKa = 3.98FF471 pKa = 4.22
Molecular weight: 51.12 kDa
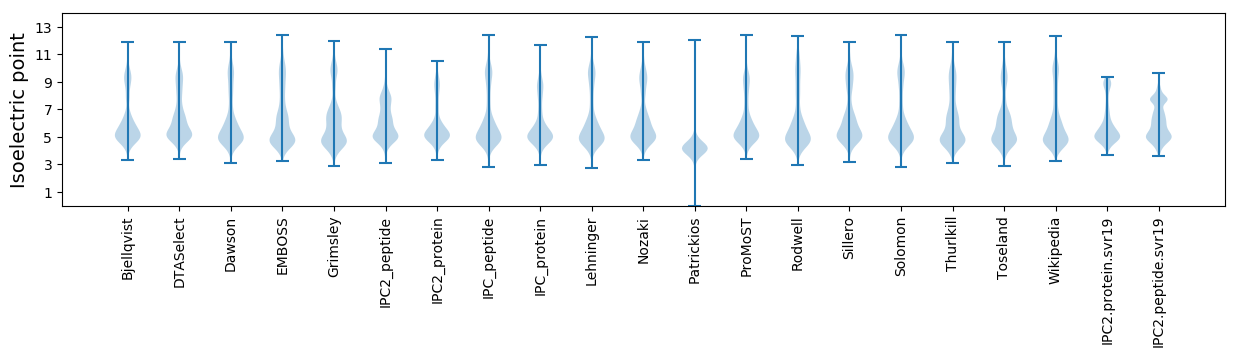
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0YDP3|A0YDP3_9GAMM Uncharacterized protein OS=marine gamma proteobacterium HTCC2143 OX=247633 GN=GP2143_10032 PE=4 SV=1
MM1 pKa = 7.39RR2 pKa = 11.84QGIGDD7 pKa = 3.7NPEE10 pKa = 4.08GQEE13 pKa = 4.05SAKK16 pKa = 10.27SGRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84MRR23 pKa = 11.84APHH26 pKa = 4.91YY27 pKa = 9.85QGYY30 pKa = 8.22PSQYY34 pKa = 8.31NTSITCRR41 pKa = 11.84VSGLPPKK48 pKa = 10.61ARR50 pKa = 11.84LKK52 pKa = 10.93KK53 pKa = 9.76LTFDD57 pKa = 4.07LGQSVRR63 pKa = 11.84HH64 pKa = 4.99VQNNVTTRR72 pKa = 11.84CTTVSFGIKK81 pKa = 10.19GPIVAAGVRR90 pKa = 11.84VMDD93 pKa = 4.27FVMLLFGYY101 pKa = 9.34FLSRR105 pKa = 11.84RR106 pKa = 11.84FYY108 pKa = 10.71FF109 pKa = 4.02
MM1 pKa = 7.39RR2 pKa = 11.84QGIGDD7 pKa = 3.7NPEE10 pKa = 4.08GQEE13 pKa = 4.05SAKK16 pKa = 10.27SGRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84MRR23 pKa = 11.84APHH26 pKa = 4.91YY27 pKa = 9.85QGYY30 pKa = 8.22PSQYY34 pKa = 8.31NTSITCRR41 pKa = 11.84VSGLPPKK48 pKa = 10.61ARR50 pKa = 11.84LKK52 pKa = 10.93KK53 pKa = 9.76LTFDD57 pKa = 4.07LGQSVRR63 pKa = 11.84HH64 pKa = 4.99VQNNVTTRR72 pKa = 11.84CTTVSFGIKK81 pKa = 10.19GPIVAAGVRR90 pKa = 11.84VMDD93 pKa = 4.27FVMLLFGYY101 pKa = 9.34FLSRR105 pKa = 11.84RR106 pKa = 11.84FYY108 pKa = 10.71FF109 pKa = 4.02
Molecular weight: 12.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1126783 |
20 |
2218 |
307.8 |
33.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.192 ± 0.04 | 1.04 ± 0.015 |
5.946 ± 0.03 | 5.968 ± 0.032 |
3.959 ± 0.027 | 7.451 ± 0.04 |
2.146 ± 0.021 | 6.431 ± 0.035 |
4.472 ± 0.03 | 10.064 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.648 ± 0.021 | 3.945 ± 0.026 |
4.184 ± 0.026 | 4.139 ± 0.033 |
5.162 ± 0.029 | 6.756 ± 0.031 |
5.307 ± 0.028 | 7.026 ± 0.034 |
1.285 ± 0.016 | 2.881 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
