
Sphaerulina musiva (strain SO2202) (Poplar stem canker fungus) (Septoria musiva)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Mycosphaerellaceae; Sphaerulina; Sphaerulina musiva
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
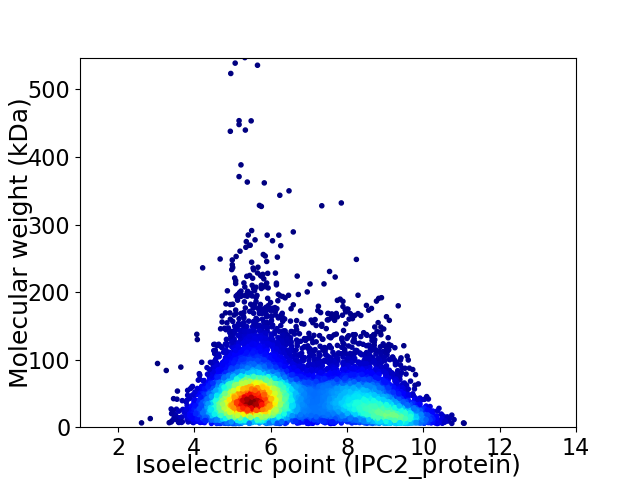
Virtual 2D-PAGE plot for 10154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M3AXA3|M3AXA3_SPHMS Uncharacterized protein OS=Sphaerulina musiva (strain SO2202) OX=692275 GN=SEPMUDRAFT_164868 PE=3 SV=1
MM1 pKa = 7.34RR2 pKa = 11.84QADD5 pKa = 3.99GVAAASLLLLLLPCILAQQITSTISIPYY33 pKa = 9.55CSTSPGLFTIYY44 pKa = 10.91GSMTSAIPSNILSSNSQTPTAGLLSSSVSSVATPLASAYY83 pKa = 9.97VDD85 pKa = 3.52QYY87 pKa = 10.23GGKK90 pKa = 8.54YY91 pKa = 7.57TVQFDD96 pKa = 3.57TGLVGSTIPASRR108 pKa = 11.84IDD110 pKa = 3.7GLRR113 pKa = 11.84KK114 pKa = 9.51RR115 pKa = 11.84QAIDD119 pKa = 3.51IGTLDD124 pKa = 3.62QTFEE128 pKa = 4.56DD129 pKa = 4.49CLEE132 pKa = 4.37SCTLTSACVAVAALDD147 pKa = 3.83STCQLFSSVRR157 pKa = 11.84EE158 pKa = 4.11TVSLVGGKK166 pKa = 8.99VAFNQEE172 pKa = 3.67RR173 pKa = 11.84FLGFSNGTILPSSMATEE190 pKa = 4.41ASSAQGMASASAISSGGFRR209 pKa = 11.84SSMPGLASMPGGSMTPLSQLPSTLAMEE236 pKa = 4.85TSTAGPLVSSFVTQDD251 pKa = 2.88ISPAGSSSDD260 pKa = 3.28IMPSAALSTNTVTTILSSGAVLSATSEE287 pKa = 4.12PLSTFSSAGDD297 pKa = 3.52PTITLQVPSSLSTEE311 pKa = 4.08SSVSISSTSSTPTDD325 pKa = 3.38QLVSTMTSDD334 pKa = 4.49GEE336 pKa = 4.07ISYY339 pKa = 8.31STDD342 pKa = 3.03LTITSSLSSLATPASASISSAASSSLALSSVASVSSSRR380 pKa = 11.84SRR382 pKa = 11.84AEE384 pKa = 3.92SSSSSSSPGPINASTFFSSPAVPTTTRR411 pKa = 11.84TTTTTTSSSSSSSSPATPTSSRR433 pKa = 11.84KK434 pKa = 9.52PPTCPDD440 pKa = 4.17DD441 pKa = 5.54DD442 pKa = 6.04GEE444 pKa = 5.67DD445 pKa = 3.85YY446 pKa = 11.1IDD448 pKa = 3.6TNGFIYY454 pKa = 10.21IITCDD459 pKa = 2.88IGFNDD464 pKa = 3.94EE465 pKa = 4.6TIQTTTVQNFVDD477 pKa = 5.98CITTCDD483 pKa = 4.27AFNEE487 pKa = 4.16LPKK490 pKa = 10.72DD491 pKa = 3.64DD492 pKa = 4.83EE493 pKa = 4.47EE494 pKa = 5.26VMGCGGVNFNSLAQTDD510 pKa = 4.03NCLLMPGAEE519 pKa = 4.02IAEE522 pKa = 4.29MAGIWSAQLSGSIAEE537 pKa = 4.24
MM1 pKa = 7.34RR2 pKa = 11.84QADD5 pKa = 3.99GVAAASLLLLLLPCILAQQITSTISIPYY33 pKa = 9.55CSTSPGLFTIYY44 pKa = 10.91GSMTSAIPSNILSSNSQTPTAGLLSSSVSSVATPLASAYY83 pKa = 9.97VDD85 pKa = 3.52QYY87 pKa = 10.23GGKK90 pKa = 8.54YY91 pKa = 7.57TVQFDD96 pKa = 3.57TGLVGSTIPASRR108 pKa = 11.84IDD110 pKa = 3.7GLRR113 pKa = 11.84KK114 pKa = 9.51RR115 pKa = 11.84QAIDD119 pKa = 3.51IGTLDD124 pKa = 3.62QTFEE128 pKa = 4.56DD129 pKa = 4.49CLEE132 pKa = 4.37SCTLTSACVAVAALDD147 pKa = 3.83STCQLFSSVRR157 pKa = 11.84EE158 pKa = 4.11TVSLVGGKK166 pKa = 8.99VAFNQEE172 pKa = 3.67RR173 pKa = 11.84FLGFSNGTILPSSMATEE190 pKa = 4.41ASSAQGMASASAISSGGFRR209 pKa = 11.84SSMPGLASMPGGSMTPLSQLPSTLAMEE236 pKa = 4.85TSTAGPLVSSFVTQDD251 pKa = 2.88ISPAGSSSDD260 pKa = 3.28IMPSAALSTNTVTTILSSGAVLSATSEE287 pKa = 4.12PLSTFSSAGDD297 pKa = 3.52PTITLQVPSSLSTEE311 pKa = 4.08SSVSISSTSSTPTDD325 pKa = 3.38QLVSTMTSDD334 pKa = 4.49GEE336 pKa = 4.07ISYY339 pKa = 8.31STDD342 pKa = 3.03LTITSSLSSLATPASASISSAASSSLALSSVASVSSSRR380 pKa = 11.84SRR382 pKa = 11.84AEE384 pKa = 3.92SSSSSSSPGPINASTFFSSPAVPTTTRR411 pKa = 11.84TTTTTTSSSSSSSSPATPTSSRR433 pKa = 11.84KK434 pKa = 9.52PPTCPDD440 pKa = 4.17DD441 pKa = 5.54DD442 pKa = 6.04GEE444 pKa = 5.67DD445 pKa = 3.85YY446 pKa = 11.1IDD448 pKa = 3.6TNGFIYY454 pKa = 10.21IITCDD459 pKa = 2.88IGFNDD464 pKa = 3.94EE465 pKa = 4.6TIQTTTVQNFVDD477 pKa = 5.98CITTCDD483 pKa = 4.27AFNEE487 pKa = 4.16LPKK490 pKa = 10.72DD491 pKa = 3.64DD492 pKa = 4.83EE493 pKa = 4.47EE494 pKa = 5.26VMGCGGVNFNSLAQTDD510 pKa = 4.03NCLLMPGAEE519 pKa = 4.02IAEE522 pKa = 4.29MAGIWSAQLSGSIAEE537 pKa = 4.24
Molecular weight: 54.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N1QH86|N1QH86_SPHMS Mitochondrial ribosomal protein S19 OS=Sphaerulina musiva (strain SO2202) OX=692275 GN=SEPMUDRAFT_72028 PE=3 SV=1
RR1 pKa = 7.12GNSRR5 pKa = 11.84IGGNSTPNRR14 pKa = 11.84NGRR17 pKa = 11.84ISGNSTPNGNSRR29 pKa = 11.84IGGNSTPNRR38 pKa = 11.84NGRR41 pKa = 11.84ISGNSTPNGNSRR53 pKa = 11.84IGGNSS58 pKa = 2.85
RR1 pKa = 7.12GNSRR5 pKa = 11.84IGGNSTPNRR14 pKa = 11.84NGRR17 pKa = 11.84ISGNSTPNGNSRR29 pKa = 11.84IGGNSTPNRR38 pKa = 11.84NGRR41 pKa = 11.84ISGNSTPNGNSRR53 pKa = 11.84IGGNSS58 pKa = 2.85
Molecular weight: 5.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4580991 |
49 |
4947 |
451.2 |
49.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.089 ± 0.021 | 1.203 ± 0.009 |
5.72 ± 0.018 | 6.216 ± 0.021 |
3.472 ± 0.015 | 6.968 ± 0.021 |
2.576 ± 0.01 | 4.621 ± 0.015 |
4.849 ± 0.022 | 8.613 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.247 ± 0.009 | 3.585 ± 0.015 |
5.949 ± 0.028 | 4.342 ± 0.018 |
6.184 ± 0.025 | 8.145 ± 0.031 |
6.113 ± 0.018 | 5.971 ± 0.016 |
1.411 ± 0.01 | 2.726 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
