
Pontivivens insulae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pontivivens
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
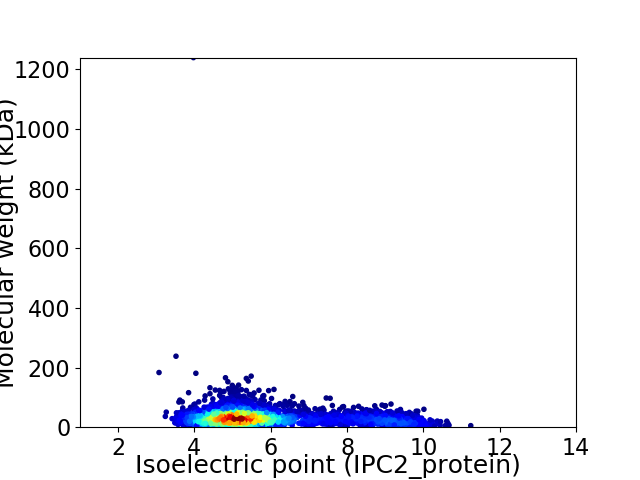
Virtual 2D-PAGE plot for 3498 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R8AF50|A0A2R8AF50_9RHOB Beta-ketodecanoyl-[acyl-carrier-protein] synthase OS=Pontivivens insulae OX=1639689 GN=POI8812_03219 PE=4 SV=1
MM1 pKa = 7.36SFSKK5 pKa = 8.03TTAIAALMTAAAALPAAAQDD25 pKa = 3.99LCGGAAANGTWIGGSDD41 pKa = 3.85EE42 pKa = 4.38TSDD45 pKa = 3.25IASGFGPFDD54 pKa = 3.24MTAQVPAGGEE64 pKa = 3.8HH65 pKa = 4.98VAIFNVSTASDD76 pKa = 3.74VRR78 pKa = 11.84VEE80 pKa = 4.11AAPVADD86 pKa = 4.21GDD88 pKa = 4.48TVIDD92 pKa = 4.4LLDD95 pKa = 3.83ASGVVILSDD104 pKa = 3.8DD105 pKa = 4.02DD106 pKa = 4.55GGGFLASRR114 pKa = 11.84GEE116 pKa = 3.81ISLDD120 pKa = 3.11PGTYY124 pKa = 10.17CLATRR129 pKa = 11.84SYY131 pKa = 11.88DD132 pKa = 4.01DD133 pKa = 3.9AALTADD139 pKa = 3.54VRR141 pKa = 11.84VGLTVHH147 pKa = 7.18DD148 pKa = 5.72PITSGGFNDD157 pKa = 3.96YY158 pKa = 11.2DD159 pKa = 3.83AVCLPEE165 pKa = 4.1TEE167 pKa = 4.31AVSFGGTNLNEE178 pKa = 4.02GLEE181 pKa = 4.32AGVAATASIGDD192 pKa = 3.63TPFYY196 pKa = 10.46RR197 pKa = 11.84FNLSEE202 pKa = 4.09PTSISFTAEE211 pKa = 3.73NQSADD216 pKa = 3.87PILRR220 pKa = 11.84IFDD223 pKa = 3.34NTGYY227 pKa = 9.66MLAEE231 pKa = 4.22NDD233 pKa = 5.1DD234 pKa = 4.01YY235 pKa = 12.03DD236 pKa = 4.17GLNSRR241 pKa = 11.84LDD243 pKa = 3.57FVQPLEE249 pKa = 4.35AGTYY253 pKa = 9.91CIGLIALSDD262 pKa = 3.66PGAPVTVTANAFSEE276 pKa = 3.98QDD278 pKa = 3.71YY279 pKa = 11.71LEE281 pKa = 4.2TLYY284 pKa = 11.49NNAEE288 pKa = 4.18TSPPLDD294 pKa = 3.0GSYY297 pKa = 10.76PVTDD301 pKa = 4.71LGLISTRR308 pKa = 11.84ARR310 pKa = 11.84ADD312 pKa = 3.14VRR314 pKa = 11.84VDD316 pKa = 5.28DD317 pKa = 5.05DD318 pKa = 5.65LIWHH322 pKa = 6.17QFQVDD327 pKa = 3.85QPGLILIEE335 pKa = 4.79GIGTGGVDD343 pKa = 4.58AMLTLFDD350 pKa = 4.75DD351 pKa = 5.05FGRR354 pKa = 11.84MITQNDD360 pKa = 4.35DD361 pKa = 3.26YY362 pKa = 11.23GQSLDD367 pKa = 3.52AQIAARR373 pKa = 11.84VQPGTYY379 pKa = 9.17VLAVGRR385 pKa = 11.84PADD388 pKa = 3.67YY389 pKa = 10.04FYY391 pKa = 11.38GEE393 pKa = 4.14PGITRR398 pKa = 11.84VGIEE402 pKa = 4.01MFVSARR408 pKa = 3.45
MM1 pKa = 7.36SFSKK5 pKa = 8.03TTAIAALMTAAAALPAAAQDD25 pKa = 3.99LCGGAAANGTWIGGSDD41 pKa = 3.85EE42 pKa = 4.38TSDD45 pKa = 3.25IASGFGPFDD54 pKa = 3.24MTAQVPAGGEE64 pKa = 3.8HH65 pKa = 4.98VAIFNVSTASDD76 pKa = 3.74VRR78 pKa = 11.84VEE80 pKa = 4.11AAPVADD86 pKa = 4.21GDD88 pKa = 4.48TVIDD92 pKa = 4.4LLDD95 pKa = 3.83ASGVVILSDD104 pKa = 3.8DD105 pKa = 4.02DD106 pKa = 4.55GGGFLASRR114 pKa = 11.84GEE116 pKa = 3.81ISLDD120 pKa = 3.11PGTYY124 pKa = 10.17CLATRR129 pKa = 11.84SYY131 pKa = 11.88DD132 pKa = 4.01DD133 pKa = 3.9AALTADD139 pKa = 3.54VRR141 pKa = 11.84VGLTVHH147 pKa = 7.18DD148 pKa = 5.72PITSGGFNDD157 pKa = 3.96YY158 pKa = 11.2DD159 pKa = 3.83AVCLPEE165 pKa = 4.1TEE167 pKa = 4.31AVSFGGTNLNEE178 pKa = 4.02GLEE181 pKa = 4.32AGVAATASIGDD192 pKa = 3.63TPFYY196 pKa = 10.46RR197 pKa = 11.84FNLSEE202 pKa = 4.09PTSISFTAEE211 pKa = 3.73NQSADD216 pKa = 3.87PILRR220 pKa = 11.84IFDD223 pKa = 3.34NTGYY227 pKa = 9.66MLAEE231 pKa = 4.22NDD233 pKa = 5.1DD234 pKa = 4.01YY235 pKa = 12.03DD236 pKa = 4.17GLNSRR241 pKa = 11.84LDD243 pKa = 3.57FVQPLEE249 pKa = 4.35AGTYY253 pKa = 9.91CIGLIALSDD262 pKa = 3.66PGAPVTVTANAFSEE276 pKa = 3.98QDD278 pKa = 3.71YY279 pKa = 11.71LEE281 pKa = 4.2TLYY284 pKa = 11.49NNAEE288 pKa = 4.18TSPPLDD294 pKa = 3.0GSYY297 pKa = 10.76PVTDD301 pKa = 4.71LGLISTRR308 pKa = 11.84ARR310 pKa = 11.84ADD312 pKa = 3.14VRR314 pKa = 11.84VDD316 pKa = 5.28DD317 pKa = 5.05DD318 pKa = 5.65LIWHH322 pKa = 6.17QFQVDD327 pKa = 3.85QPGLILIEE335 pKa = 4.79GIGTGGVDD343 pKa = 4.58AMLTLFDD350 pKa = 4.75DD351 pKa = 5.05FGRR354 pKa = 11.84MITQNDD360 pKa = 4.35DD361 pKa = 3.26YY362 pKa = 11.23GQSLDD367 pKa = 3.52AQIAARR373 pKa = 11.84VQPGTYY379 pKa = 9.17VLAVGRR385 pKa = 11.84PADD388 pKa = 3.67YY389 pKa = 10.04FYY391 pKa = 11.38GEE393 pKa = 4.14PGITRR398 pKa = 11.84VGIEE402 pKa = 4.01MFVSARR408 pKa = 3.45
Molecular weight: 42.66 kDa
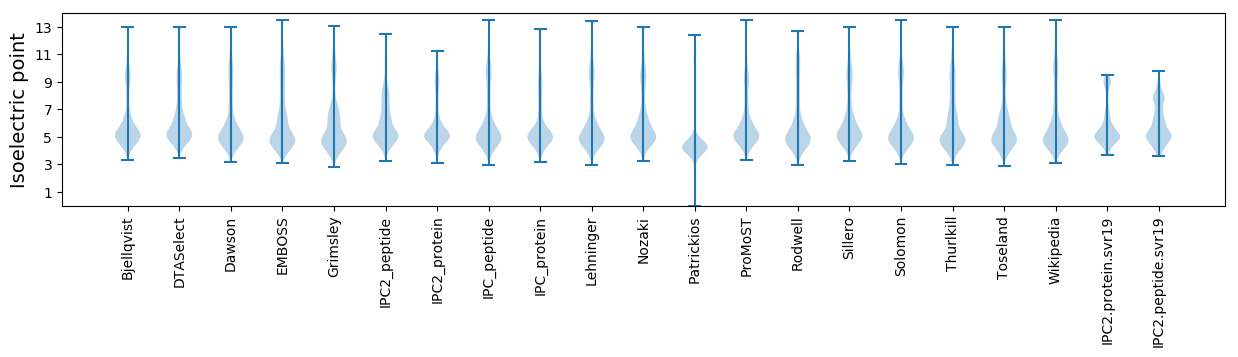
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R8AFQ0|A0A2R8AFQ0_9RHOB Uncharacterized protein OS=Pontivivens insulae OX=1639689 GN=POI8812_03424 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84KK29 pKa = 9.03ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84KK29 pKa = 9.03ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1084088 |
29 |
11983 |
309.9 |
33.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.497 ± 0.06 | 0.895 ± 0.016 |
6.235 ± 0.051 | 6.037 ± 0.044 |
3.718 ± 0.027 | 8.665 ± 0.051 |
2.04 ± 0.026 | 5.273 ± 0.027 |
2.734 ± 0.037 | 10.021 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.792 ± 0.029 | 2.598 ± 0.027 |
4.958 ± 0.028 | 3.265 ± 0.023 |
6.686 ± 0.055 | 5.173 ± 0.032 |
5.541 ± 0.05 | 7.306 ± 0.036 |
1.413 ± 0.02 | 2.154 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
