
Ceratobasidium theobromae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Cantharellales; Ceratobasidiaceae; Ceratobasidium
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
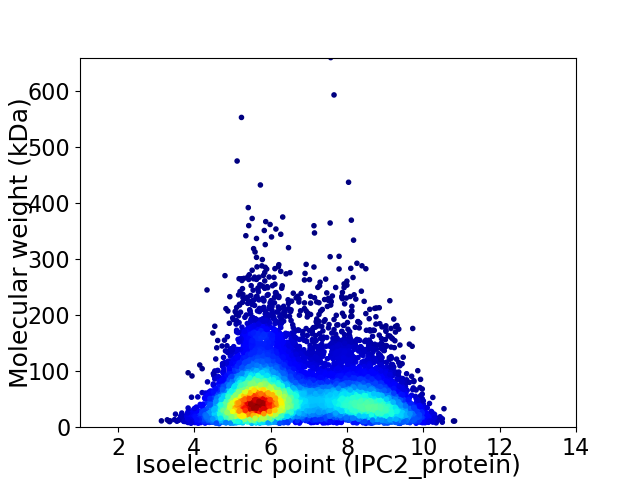
Virtual 2D-PAGE plot for 9256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N5QEB7|A0A5N5QEB7_9AGAM Mitogen-activated protein kinase OS=Ceratobasidium theobromae OX=1582974 GN=CTheo_6759 PE=3 SV=1
MM1 pKa = 7.58GISYY5 pKa = 10.41YY6 pKa = 10.78DD7 pKa = 3.7EE8 pKa = 5.76IEE10 pKa = 4.5IEE12 pKa = 4.19DD13 pKa = 4.18MVWDD17 pKa = 3.77EE18 pKa = 4.4AKK20 pKa = 10.17RR21 pKa = 11.84VYY23 pKa = 10.17HH24 pKa = 5.7YY25 pKa = 8.21PCPCGDD31 pKa = 3.19RR32 pKa = 11.84FEE34 pKa = 4.61ISRR37 pKa = 11.84EE38 pKa = 3.64QLANYY43 pKa = 9.4EE44 pKa = 4.92DD45 pKa = 4.61IATCPSCSLIIRR57 pKa = 11.84VIYY60 pKa = 10.39DD61 pKa = 3.4PLDD64 pKa = 4.11FEE66 pKa = 6.0DD67 pKa = 5.03EE68 pKa = 4.59EE69 pKa = 5.4PDD71 pKa = 4.08EE72 pKa = 5.69DD73 pKa = 5.0DD74 pKa = 4.56PDD76 pKa = 5.77PIEE79 pKa = 4.48EE80 pKa = 4.37EE81 pKa = 4.23EE82 pKa = 4.94SSGSDD87 pKa = 4.04DD88 pKa = 4.36GGKK91 pKa = 9.71EE92 pKa = 3.62ADD94 pKa = 3.43KK95 pKa = 11.03VQNAMEE101 pKa = 4.11NLALNEE107 pKa = 3.96EE108 pKa = 4.48PLAVTLVEE116 pKa = 4.43TRR118 pKa = 11.84AA119 pKa = 3.38
MM1 pKa = 7.58GISYY5 pKa = 10.41YY6 pKa = 10.78DD7 pKa = 3.7EE8 pKa = 5.76IEE10 pKa = 4.5IEE12 pKa = 4.19DD13 pKa = 4.18MVWDD17 pKa = 3.77EE18 pKa = 4.4AKK20 pKa = 10.17RR21 pKa = 11.84VYY23 pKa = 10.17HH24 pKa = 5.7YY25 pKa = 8.21PCPCGDD31 pKa = 3.19RR32 pKa = 11.84FEE34 pKa = 4.61ISRR37 pKa = 11.84EE38 pKa = 3.64QLANYY43 pKa = 9.4EE44 pKa = 4.92DD45 pKa = 4.61IATCPSCSLIIRR57 pKa = 11.84VIYY60 pKa = 10.39DD61 pKa = 3.4PLDD64 pKa = 4.11FEE66 pKa = 6.0DD67 pKa = 5.03EE68 pKa = 4.59EE69 pKa = 5.4PDD71 pKa = 4.08EE72 pKa = 5.69DD73 pKa = 5.0DD74 pKa = 4.56PDD76 pKa = 5.77PIEE79 pKa = 4.48EE80 pKa = 4.37EE81 pKa = 4.23EE82 pKa = 4.94SSGSDD87 pKa = 4.04DD88 pKa = 4.36GGKK91 pKa = 9.71EE92 pKa = 3.62ADD94 pKa = 3.43KK95 pKa = 11.03VQNAMEE101 pKa = 4.11NLALNEE107 pKa = 3.96EE108 pKa = 4.48PLAVTLVEE116 pKa = 4.43TRR118 pKa = 11.84AA119 pKa = 3.38
Molecular weight: 13.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N5QQ55|A0A5N5QQ55_9AGAM G protein coupled receptor OS=Ceratobasidium theobromae OX=1582974 GN=CTheo_3289 PE=4 SV=1
MM1 pKa = 7.36SLSPPQPLRR10 pKa = 11.84ARR12 pKa = 11.84ASSIVQPLTIDD23 pKa = 3.26SLRR26 pKa = 11.84RR27 pKa = 11.84TTGLATPPASPFSSVFTRR45 pKa = 11.84PPRR48 pKa = 11.84QHH50 pKa = 6.7LPSPALSRR58 pKa = 11.84TPSPPPRR65 pKa = 11.84EE66 pKa = 3.51THH68 pKa = 6.73APVLRR73 pKa = 11.84LAAGFALSAVPILMALSTLPVALRR97 pKa = 11.84MPRR100 pKa = 11.84TLADD104 pKa = 3.04IALLGRR110 pKa = 11.84DD111 pKa = 3.77LKK113 pKa = 11.13AYY115 pKa = 9.14AASAPGMAHH124 pKa = 6.15VMAVMCCLAVWEE136 pKa = 4.92HH137 pKa = 6.2AWSIPGSVVLNVLAGALVSPLWATLLMTLLTSLGSLAASLLAAPLEE183 pKa = 4.75PIVQRR188 pKa = 11.84IAPKK192 pKa = 10.49ALDD195 pKa = 3.68VVASALQGDD204 pKa = 3.8PRR206 pKa = 11.84AAKK209 pKa = 7.9RR210 pKa = 11.84TPTWVRR216 pKa = 11.84IVVMRR221 pKa = 11.84LVGVVPWSAINIACGVCRR239 pKa = 11.84VPFVDD244 pKa = 3.98CAIGAFIGALPWTAVTCQTLALGSSSDD271 pKa = 3.63PKK273 pKa = 10.41TLSSLLSSPAIILKK287 pKa = 10.36LVLLSLLSLGPVLCRR302 pKa = 11.84DD303 pKa = 3.71KK304 pKa = 11.19LTDD307 pKa = 3.63ALAATDD313 pKa = 4.23EE314 pKa = 4.39KK315 pKa = 11.46APLAPAPMWKK325 pKa = 8.53VWHH328 pKa = 6.24WRR330 pKa = 11.84RR331 pKa = 11.84SSHH334 pKa = 5.99GLPRR338 pKa = 11.84PKK340 pKa = 10.47SPEE343 pKa = 3.61RR344 pKa = 11.84TMEE347 pKa = 3.9EE348 pKa = 4.04VLDD351 pKa = 4.18EE352 pKa = 4.17NLIFAMSPHH361 pKa = 7.02ANHH364 pKa = 5.57VMYY367 pKa = 9.99PVLLFCSGGIQSSSLVYY384 pKa = 10.57NYY386 pKa = 10.2LIQFGLGRR394 pKa = 11.84LEE396 pKa = 4.64PGPLGIPRR404 pKa = 11.84STHH407 pKa = 6.24PNTGRR412 pKa = 11.84HH413 pKa = 5.09PAYY416 pKa = 10.25ALSQFSFQHH425 pKa = 5.85PPVRR429 pKa = 11.84PHH431 pKa = 6.82LARR434 pKa = 11.84RR435 pKa = 11.84SHH437 pKa = 6.91FDD439 pKa = 3.06VTRR442 pKa = 11.84TTWLAYY448 pKa = 9.54KK449 pKa = 10.01FASKK453 pKa = 10.53GPHH456 pKa = 6.33IFLCPASAPRR466 pKa = 11.84VSRR469 pKa = 11.84PSLRR473 pKa = 3.52
MM1 pKa = 7.36SLSPPQPLRR10 pKa = 11.84ARR12 pKa = 11.84ASSIVQPLTIDD23 pKa = 3.26SLRR26 pKa = 11.84RR27 pKa = 11.84TTGLATPPASPFSSVFTRR45 pKa = 11.84PPRR48 pKa = 11.84QHH50 pKa = 6.7LPSPALSRR58 pKa = 11.84TPSPPPRR65 pKa = 11.84EE66 pKa = 3.51THH68 pKa = 6.73APVLRR73 pKa = 11.84LAAGFALSAVPILMALSTLPVALRR97 pKa = 11.84MPRR100 pKa = 11.84TLADD104 pKa = 3.04IALLGRR110 pKa = 11.84DD111 pKa = 3.77LKK113 pKa = 11.13AYY115 pKa = 9.14AASAPGMAHH124 pKa = 6.15VMAVMCCLAVWEE136 pKa = 4.92HH137 pKa = 6.2AWSIPGSVVLNVLAGALVSPLWATLLMTLLTSLGSLAASLLAAPLEE183 pKa = 4.75PIVQRR188 pKa = 11.84IAPKK192 pKa = 10.49ALDD195 pKa = 3.68VVASALQGDD204 pKa = 3.8PRR206 pKa = 11.84AAKK209 pKa = 7.9RR210 pKa = 11.84TPTWVRR216 pKa = 11.84IVVMRR221 pKa = 11.84LVGVVPWSAINIACGVCRR239 pKa = 11.84VPFVDD244 pKa = 3.98CAIGAFIGALPWTAVTCQTLALGSSSDD271 pKa = 3.63PKK273 pKa = 10.41TLSSLLSSPAIILKK287 pKa = 10.36LVLLSLLSLGPVLCRR302 pKa = 11.84DD303 pKa = 3.71KK304 pKa = 11.19LTDD307 pKa = 3.63ALAATDD313 pKa = 4.23EE314 pKa = 4.39KK315 pKa = 11.46APLAPAPMWKK325 pKa = 8.53VWHH328 pKa = 6.24WRR330 pKa = 11.84RR331 pKa = 11.84SSHH334 pKa = 5.99GLPRR338 pKa = 11.84PKK340 pKa = 10.47SPEE343 pKa = 3.61RR344 pKa = 11.84TMEE347 pKa = 3.9EE348 pKa = 4.04VLDD351 pKa = 4.18EE352 pKa = 4.17NLIFAMSPHH361 pKa = 7.02ANHH364 pKa = 5.57VMYY367 pKa = 9.99PVLLFCSGGIQSSSLVYY384 pKa = 10.57NYY386 pKa = 10.2LIQFGLGRR394 pKa = 11.84LEE396 pKa = 4.64PGPLGIPRR404 pKa = 11.84STHH407 pKa = 6.24PNTGRR412 pKa = 11.84HH413 pKa = 5.09PAYY416 pKa = 10.25ALSQFSFQHH425 pKa = 5.85PPVRR429 pKa = 11.84PHH431 pKa = 6.82LARR434 pKa = 11.84RR435 pKa = 11.84SHH437 pKa = 6.91FDD439 pKa = 3.06VTRR442 pKa = 11.84TTWLAYY448 pKa = 9.54KK449 pKa = 10.01FASKK453 pKa = 10.53GPHH456 pKa = 6.33IFLCPASAPRR466 pKa = 11.84VSRR469 pKa = 11.84PSLRR473 pKa = 3.52
Molecular weight: 50.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5421178 |
66 |
6167 |
585.7 |
64.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.943 ± 0.021 | 1.234 ± 0.008 |
5.418 ± 0.016 | 5.688 ± 0.025 |
3.501 ± 0.014 | 6.899 ± 0.025 |
2.513 ± 0.01 | 4.944 ± 0.016 |
4.322 ± 0.02 | 8.998 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.027 ± 0.009 | 3.502 ± 0.013 |
6.807 ± 0.031 | 3.735 ± 0.013 |
6.29 ± 0.021 | 8.668 ± 0.027 |
6.181 ± 0.016 | 6.337 ± 0.015 |
1.466 ± 0.009 | 2.53 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
