
Lamium leaf distortion virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Caulimovirus
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
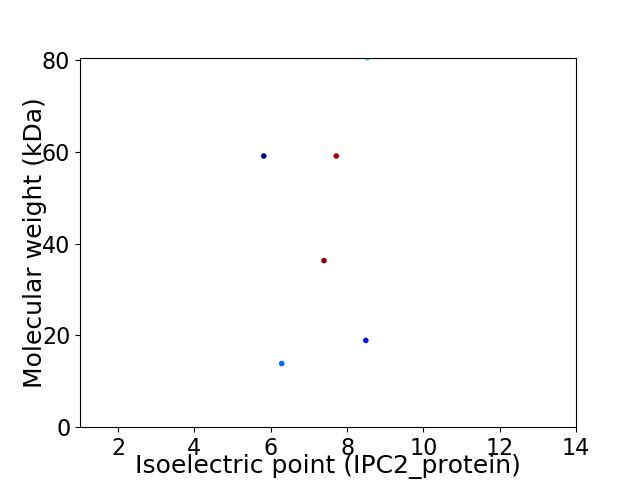
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2CXY5|B2CXY5_9VIRU Aspartic protease OS=Lamium leaf distortion virus OX=515320 PE=3 SV=1
MM1 pKa = 7.49SFYY4 pKa = 11.2EE5 pKa = 5.51LIDD8 pKa = 3.46KK9 pKa = 8.65QFWEE13 pKa = 4.23TLEE16 pKa = 4.31TQSEE20 pKa = 4.11LDD22 pKa = 3.36EE23 pKa = 4.51RR24 pKa = 11.84LNTLSRR30 pKa = 11.84LVNQGEE36 pKa = 4.11QATNEE41 pKa = 3.95EE42 pKa = 4.27LQGNEE47 pKa = 3.91TLRR50 pKa = 11.84SFLNLDD56 pKa = 4.04NICEE60 pKa = 4.09RR61 pKa = 11.84FEE63 pKa = 4.03TLNVIEE69 pKa = 4.93EE70 pKa = 4.22SSEE73 pKa = 4.28DD74 pKa = 3.88SSIDD78 pKa = 3.3NNDD81 pKa = 3.36PVDD84 pKa = 3.88YY85 pKa = 10.39SDD87 pKa = 4.02SSSEE91 pKa = 4.17EE92 pKa = 3.94EE93 pKa = 3.95EE94 pKa = 4.03QNTNKK99 pKa = 10.09RR100 pKa = 11.84FFMEE104 pKa = 4.46KK105 pKa = 10.87GEE107 pKa = 4.2GSQNKK112 pKa = 9.0KK113 pKa = 10.82DD114 pKa = 3.72KK115 pKa = 10.44DD116 pKa = 3.75PKK118 pKa = 10.01VEE120 pKa = 4.03YY121 pKa = 10.17PNVEE125 pKa = 4.0NLQSEE130 pKa = 4.75DD131 pKa = 3.25NRR133 pKa = 11.84RR134 pKa = 11.84TPFNNNKK141 pKa = 9.69RR142 pKa = 11.84KK143 pKa = 10.12SPEE146 pKa = 3.52QARR149 pKa = 11.84YY150 pKa = 7.61PVKK153 pKa = 10.48PKK155 pKa = 10.65FEE157 pKa = 4.41VVSRR161 pKa = 11.84TLGFLDD167 pKa = 4.79IDD169 pKa = 4.34CQDD172 pKa = 3.3HH173 pKa = 5.91EE174 pKa = 4.56VRR176 pKa = 11.84KK177 pKa = 10.55KK178 pKa = 10.85NIDD181 pKa = 2.9AWASEE186 pKa = 4.15LSLIVQTNPDD196 pKa = 3.57AYY198 pKa = 10.62DD199 pKa = 3.51NADD202 pKa = 3.46AVILLAEE209 pKa = 4.68HH210 pKa = 6.86KK211 pKa = 10.79ALGNAKK217 pKa = 9.57EE218 pKa = 4.47LIKK221 pKa = 10.16GTYY224 pKa = 7.33WNKK227 pKa = 10.65DD228 pKa = 3.47NPPMYY233 pKa = 9.66MLNQIFDD240 pKa = 3.79SLYY243 pKa = 11.34SMFLGLDD250 pKa = 3.35YY251 pKa = 11.16TDD253 pKa = 5.61AISKK257 pKa = 10.29LLEE260 pKa = 3.94TNKK263 pKa = 9.97QKK265 pKa = 11.01EE266 pKa = 4.02NARR269 pKa = 11.84IRR271 pKa = 11.84LANLKK276 pKa = 9.81LCNICSIDD284 pKa = 3.87EE285 pKa = 4.29FNCAYY290 pKa = 10.18EE291 pKa = 4.22EE292 pKa = 4.26EE293 pKa = 4.42FFKK296 pKa = 11.18LNQNEE301 pKa = 4.15YY302 pKa = 9.46PKK304 pKa = 10.9YY305 pKa = 10.27ISDD308 pKa = 4.33YY309 pKa = 9.85LLKK312 pKa = 10.53IPVVGLAAKK321 pKa = 10.13EE322 pKa = 4.28KK323 pKa = 10.13FDD325 pKa = 3.76KK326 pKa = 8.95TTGTLAYY333 pKa = 10.4SIGHH337 pKa = 6.0AEE339 pKa = 3.82KK340 pKa = 10.54LVRR343 pKa = 11.84EE344 pKa = 4.85EE345 pKa = 3.53ITKK348 pKa = 9.89ICEE351 pKa = 3.98LSQQQKK357 pKa = 10.19KK358 pKa = 9.82LKK360 pKa = 10.15KK361 pKa = 9.84ISKK364 pKa = 9.14KK365 pKa = 10.05CCNKK369 pKa = 9.77IANKK373 pKa = 8.93STDD376 pKa = 2.95IGCYY380 pKa = 5.09TTKK383 pKa = 10.43SYY385 pKa = 11.09KK386 pKa = 9.89KK387 pKa = 9.91KK388 pKa = 10.17KK389 pKa = 9.02KK390 pKa = 10.17KK391 pKa = 10.08KK392 pKa = 7.41FKK394 pKa = 10.49KK395 pKa = 10.03YY396 pKa = 10.13VFSKK400 pKa = 10.49RR401 pKa = 11.84KK402 pKa = 8.95RR403 pKa = 11.84KK404 pKa = 9.58KK405 pKa = 9.9FRR407 pKa = 11.84PGKK410 pKa = 9.67YY411 pKa = 8.43FQKK414 pKa = 10.77KK415 pKa = 9.12KK416 pKa = 10.61FPTSGEE422 pKa = 4.19GKK424 pKa = 10.14KK425 pKa = 9.89PCPKK429 pKa = 10.55GKK431 pKa = 8.6TNCRR435 pKa = 11.84CWICNVEE442 pKa = 3.6GHH444 pKa = 6.06YY445 pKa = 11.17ANKK448 pKa = 10.24CPNKK452 pKa = 9.64QKK454 pKa = 10.78YY455 pKa = 8.2GEE457 pKa = 4.21KK458 pKa = 10.6VKK460 pKa = 10.47ILRR463 pKa = 11.84KK464 pKa = 9.74ADD466 pKa = 3.76LQDD469 pKa = 3.97LEE471 pKa = 4.8PIEE474 pKa = 5.77DD475 pKa = 3.55IFEE478 pKa = 4.37EE479 pKa = 4.31SLDD482 pKa = 3.72VYY484 pKa = 10.18IAEE487 pKa = 4.92IIEE490 pKa = 4.08FDD492 pKa = 3.94PDD494 pKa = 3.24LSYY497 pKa = 11.34QEE499 pKa = 5.15SSDD502 pKa = 3.74EE503 pKa = 4.15SSEE506 pKa = 4.15EE507 pKa = 3.82SDD509 pKa = 3.38
MM1 pKa = 7.49SFYY4 pKa = 11.2EE5 pKa = 5.51LIDD8 pKa = 3.46KK9 pKa = 8.65QFWEE13 pKa = 4.23TLEE16 pKa = 4.31TQSEE20 pKa = 4.11LDD22 pKa = 3.36EE23 pKa = 4.51RR24 pKa = 11.84LNTLSRR30 pKa = 11.84LVNQGEE36 pKa = 4.11QATNEE41 pKa = 3.95EE42 pKa = 4.27LQGNEE47 pKa = 3.91TLRR50 pKa = 11.84SFLNLDD56 pKa = 4.04NICEE60 pKa = 4.09RR61 pKa = 11.84FEE63 pKa = 4.03TLNVIEE69 pKa = 4.93EE70 pKa = 4.22SSEE73 pKa = 4.28DD74 pKa = 3.88SSIDD78 pKa = 3.3NNDD81 pKa = 3.36PVDD84 pKa = 3.88YY85 pKa = 10.39SDD87 pKa = 4.02SSSEE91 pKa = 4.17EE92 pKa = 3.94EE93 pKa = 3.95EE94 pKa = 4.03QNTNKK99 pKa = 10.09RR100 pKa = 11.84FFMEE104 pKa = 4.46KK105 pKa = 10.87GEE107 pKa = 4.2GSQNKK112 pKa = 9.0KK113 pKa = 10.82DD114 pKa = 3.72KK115 pKa = 10.44DD116 pKa = 3.75PKK118 pKa = 10.01VEE120 pKa = 4.03YY121 pKa = 10.17PNVEE125 pKa = 4.0NLQSEE130 pKa = 4.75DD131 pKa = 3.25NRR133 pKa = 11.84RR134 pKa = 11.84TPFNNNKK141 pKa = 9.69RR142 pKa = 11.84KK143 pKa = 10.12SPEE146 pKa = 3.52QARR149 pKa = 11.84YY150 pKa = 7.61PVKK153 pKa = 10.48PKK155 pKa = 10.65FEE157 pKa = 4.41VVSRR161 pKa = 11.84TLGFLDD167 pKa = 4.79IDD169 pKa = 4.34CQDD172 pKa = 3.3HH173 pKa = 5.91EE174 pKa = 4.56VRR176 pKa = 11.84KK177 pKa = 10.55KK178 pKa = 10.85NIDD181 pKa = 2.9AWASEE186 pKa = 4.15LSLIVQTNPDD196 pKa = 3.57AYY198 pKa = 10.62DD199 pKa = 3.51NADD202 pKa = 3.46AVILLAEE209 pKa = 4.68HH210 pKa = 6.86KK211 pKa = 10.79ALGNAKK217 pKa = 9.57EE218 pKa = 4.47LIKK221 pKa = 10.16GTYY224 pKa = 7.33WNKK227 pKa = 10.65DD228 pKa = 3.47NPPMYY233 pKa = 9.66MLNQIFDD240 pKa = 3.79SLYY243 pKa = 11.34SMFLGLDD250 pKa = 3.35YY251 pKa = 11.16TDD253 pKa = 5.61AISKK257 pKa = 10.29LLEE260 pKa = 3.94TNKK263 pKa = 9.97QKK265 pKa = 11.01EE266 pKa = 4.02NARR269 pKa = 11.84IRR271 pKa = 11.84LANLKK276 pKa = 9.81LCNICSIDD284 pKa = 3.87EE285 pKa = 4.29FNCAYY290 pKa = 10.18EE291 pKa = 4.22EE292 pKa = 4.26EE293 pKa = 4.42FFKK296 pKa = 11.18LNQNEE301 pKa = 4.15YY302 pKa = 9.46PKK304 pKa = 10.9YY305 pKa = 10.27ISDD308 pKa = 4.33YY309 pKa = 9.85LLKK312 pKa = 10.53IPVVGLAAKK321 pKa = 10.13EE322 pKa = 4.28KK323 pKa = 10.13FDD325 pKa = 3.76KK326 pKa = 8.95TTGTLAYY333 pKa = 10.4SIGHH337 pKa = 6.0AEE339 pKa = 3.82KK340 pKa = 10.54LVRR343 pKa = 11.84EE344 pKa = 4.85EE345 pKa = 3.53ITKK348 pKa = 9.89ICEE351 pKa = 3.98LSQQQKK357 pKa = 10.19KK358 pKa = 9.82LKK360 pKa = 10.15KK361 pKa = 9.84ISKK364 pKa = 9.14KK365 pKa = 10.05CCNKK369 pKa = 9.77IANKK373 pKa = 8.93STDD376 pKa = 2.95IGCYY380 pKa = 5.09TTKK383 pKa = 10.43SYY385 pKa = 11.09KK386 pKa = 9.89KK387 pKa = 9.91KK388 pKa = 10.17KK389 pKa = 9.02KK390 pKa = 10.17KK391 pKa = 10.08KK392 pKa = 7.41FKK394 pKa = 10.49KK395 pKa = 10.03YY396 pKa = 10.13VFSKK400 pKa = 10.49RR401 pKa = 11.84KK402 pKa = 8.95RR403 pKa = 11.84KK404 pKa = 9.58KK405 pKa = 9.9FRR407 pKa = 11.84PGKK410 pKa = 9.67YY411 pKa = 8.43FQKK414 pKa = 10.77KK415 pKa = 9.12KK416 pKa = 10.61FPTSGEE422 pKa = 4.19GKK424 pKa = 10.14KK425 pKa = 9.89PCPKK429 pKa = 10.55GKK431 pKa = 8.6TNCRR435 pKa = 11.84CWICNVEE442 pKa = 3.6GHH444 pKa = 6.06YY445 pKa = 11.17ANKK448 pKa = 10.24CPNKK452 pKa = 9.64QKK454 pKa = 10.78YY455 pKa = 8.2GEE457 pKa = 4.21KK458 pKa = 10.6VKK460 pKa = 10.47ILRR463 pKa = 11.84KK464 pKa = 9.74ADD466 pKa = 3.76LQDD469 pKa = 3.97LEE471 pKa = 4.8PIEE474 pKa = 5.77DD475 pKa = 3.55IFEE478 pKa = 4.37EE479 pKa = 4.31SLDD482 pKa = 3.72VYY484 pKa = 10.18IAEE487 pKa = 4.92IIEE490 pKa = 4.08FDD492 pKa = 3.94PDD494 pKa = 3.24LSYY497 pKa = 11.34QEE499 pKa = 5.15SSDD502 pKa = 3.74EE503 pKa = 4.15SSEE506 pKa = 4.15EE507 pKa = 3.82SDD509 pKa = 3.38
Molecular weight: 59.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2CXY3|B2CXY3_9VIRU Protein 3 OS=Lamium leaf distortion virus OX=515320 PE=3 SV=1
MM1 pKa = 7.6SITSKK6 pKa = 10.2SHH8 pKa = 5.85IYY10 pKa = 10.44KK11 pKa = 10.38KK12 pKa = 10.03GTTIIPLKK20 pKa = 10.49NLAINTDD27 pKa = 2.98GKK29 pKa = 11.11KK30 pKa = 10.32YY31 pKa = 10.72VFNSLKK37 pKa = 10.62PNIQSVVNHH46 pKa = 6.23CNNLNEE52 pKa = 4.13VCGRR56 pKa = 11.84ILLGIWKK63 pKa = 8.92LCSYY67 pKa = 10.58FGLSKK72 pKa = 10.9DD73 pKa = 3.27PSEE76 pKa = 4.14PHH78 pKa = 6.15SKK80 pKa = 10.46NPSVYY85 pKa = 10.7DD86 pKa = 3.41HH87 pKa = 7.27AKK89 pKa = 10.03TIFKK93 pKa = 10.62SGGVDD98 pKa = 2.85HH99 pKa = 6.9SVILRR104 pKa = 11.84EE105 pKa = 3.91IKK107 pKa = 10.64SLIEE111 pKa = 3.81TQQTKK116 pKa = 10.53NKK118 pKa = 9.77NLEE121 pKa = 4.02NKK123 pKa = 9.49IDD125 pKa = 3.86NLEE128 pKa = 4.03KK129 pKa = 10.59SIKK132 pKa = 10.06DD133 pKa = 3.41LSHH136 pKa = 7.11KK137 pKa = 10.31IEE139 pKa = 4.67PEE141 pKa = 3.81PLTHH145 pKa = 6.97EE146 pKa = 4.66KK147 pKa = 10.45IKK149 pKa = 11.05DD150 pKa = 3.39FSNALKK156 pKa = 10.81AIDD159 pKa = 4.99DD160 pKa = 3.84KK161 pKa = 11.42LKK163 pKa = 11.15NVIGEE168 pKa = 4.08
MM1 pKa = 7.6SITSKK6 pKa = 10.2SHH8 pKa = 5.85IYY10 pKa = 10.44KK11 pKa = 10.38KK12 pKa = 10.03GTTIIPLKK20 pKa = 10.49NLAINTDD27 pKa = 2.98GKK29 pKa = 11.11KK30 pKa = 10.32YY31 pKa = 10.72VFNSLKK37 pKa = 10.62PNIQSVVNHH46 pKa = 6.23CNNLNEE52 pKa = 4.13VCGRR56 pKa = 11.84ILLGIWKK63 pKa = 8.92LCSYY67 pKa = 10.58FGLSKK72 pKa = 10.9DD73 pKa = 3.27PSEE76 pKa = 4.14PHH78 pKa = 6.15SKK80 pKa = 10.46NPSVYY85 pKa = 10.7DD86 pKa = 3.41HH87 pKa = 7.27AKK89 pKa = 10.03TIFKK93 pKa = 10.62SGGVDD98 pKa = 2.85HH99 pKa = 6.9SVILRR104 pKa = 11.84EE105 pKa = 3.91IKK107 pKa = 10.64SLIEE111 pKa = 3.81TQQTKK116 pKa = 10.53NKK118 pKa = 9.77NLEE121 pKa = 4.02NKK123 pKa = 9.49IDD125 pKa = 3.86NLEE128 pKa = 4.03KK129 pKa = 10.59SIKK132 pKa = 10.06DD133 pKa = 3.41LSHH136 pKa = 7.11KK137 pKa = 10.31IEE139 pKa = 4.67PEE141 pKa = 3.81PLTHH145 pKa = 6.97EE146 pKa = 4.66KK147 pKa = 10.45IKK149 pKa = 11.05DD150 pKa = 3.39FSNALKK156 pKa = 10.81AIDD159 pKa = 4.99DD160 pKa = 3.84KK161 pKa = 11.42LKK163 pKa = 11.15NVIGEE168 pKa = 4.08
Molecular weight: 18.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2335 |
122 |
696 |
389.2 |
44.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.411 ± 0.242 | 1.97 ± 0.261 |
5.396 ± 0.432 | 7.923 ± 0.726 |
4.368 ± 0.306 | 3.94 ± 0.22 |
2.184 ± 0.401 | 7.666 ± 0.811 |
11.221 ± 0.673 | 8.822 ± 0.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.627 ± 0.274 | 6.51 ± 0.859 |
4.411 ± 0.66 | 4.069 ± 0.3 |
3.683 ± 0.402 | 8.223 ± 0.834 |
4.668 ± 0.172 | 4.668 ± 0.35 |
0.857 ± 0.172 | 3.383 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
