
Actinidia chlorotic ringspot-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus; Actinidia chlorotic ringspot-associated emaravirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
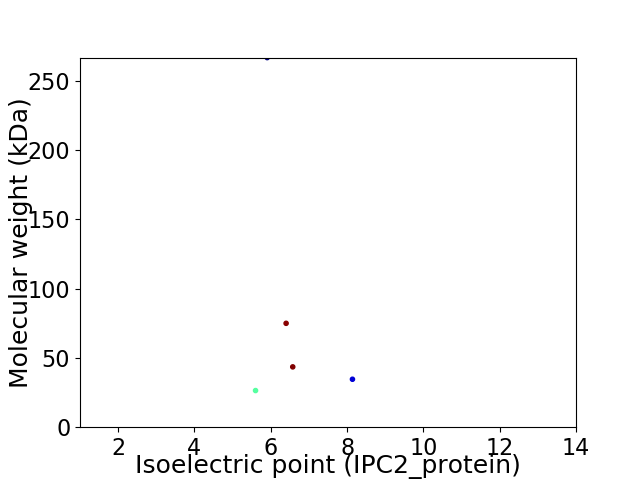
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
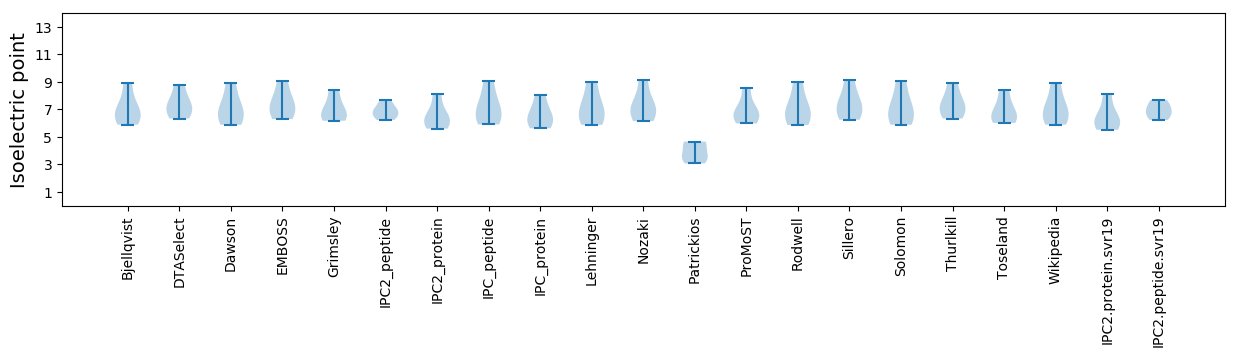
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4CCN7|A0A0U4CCN7_9VIRU Replicase OS=Actinidia chlorotic ringspot-associated virus OX=1776763 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 5.56LFKK5 pKa = 10.56TYY7 pKa = 10.64LSAFSWMYY15 pKa = 9.87EE16 pKa = 3.99QIFHH20 pKa = 7.21IISEE24 pKa = 4.46EE25 pKa = 4.14FPSKK29 pKa = 11.07YY30 pKa = 10.27YY31 pKa = 10.66DD32 pKa = 3.31NCCIIHH38 pKa = 6.9EE39 pKa = 4.57YY40 pKa = 10.42YY41 pKa = 10.12MSGTIIYY48 pKa = 9.55EE49 pKa = 4.05DD50 pKa = 4.06LFFDD54 pKa = 4.84GPHH57 pKa = 5.76KK58 pKa = 10.54RR59 pKa = 11.84IYY61 pKa = 10.85DD62 pKa = 3.66DD63 pKa = 4.17VINAGSAAYY72 pKa = 9.6YY73 pKa = 10.39RR74 pKa = 11.84KK75 pKa = 9.54FQALCFMVLGHH86 pKa = 6.9LEE88 pKa = 4.12PLFKK92 pKa = 10.81KK93 pKa = 10.14HH94 pKa = 6.31GDD96 pKa = 3.63KK97 pKa = 11.17LDD99 pKa = 3.66GLQIRR104 pKa = 11.84DD105 pKa = 3.83YY106 pKa = 11.19SSHH109 pKa = 7.24LDD111 pKa = 3.21QGLCYY116 pKa = 9.48IRR118 pKa = 11.84HH119 pKa = 6.19IFRR122 pKa = 11.84GYY124 pKa = 10.9LMEE127 pKa = 4.7RR128 pKa = 11.84MNFEE132 pKa = 4.01MANQYY137 pKa = 10.39ALKK140 pKa = 10.69SFDD143 pKa = 3.25SKK145 pKa = 10.61MVYY148 pKa = 8.25GTPSFEE154 pKa = 4.63FYY156 pKa = 10.58SKK158 pKa = 10.98SIMYY162 pKa = 9.43LQLKK166 pKa = 9.4HH167 pKa = 6.63ISTISDD173 pKa = 3.53SLKK176 pKa = 11.1DD177 pKa = 3.54GLKK180 pKa = 10.27SLKK183 pKa = 10.95SNDD186 pKa = 4.0LSDD189 pKa = 5.72DD190 pKa = 3.81DD191 pKa = 5.64DD192 pKa = 4.65EE193 pKa = 6.55SGEE196 pKa = 4.05ILKK199 pKa = 10.74KK200 pKa = 10.79LLLDD204 pKa = 3.92NTDD207 pKa = 3.25QTASTSCKK215 pKa = 9.81LKK217 pKa = 10.35RR218 pKa = 11.84QISIISIKK226 pKa = 10.67
MM1 pKa = 7.58EE2 pKa = 5.56LFKK5 pKa = 10.56TYY7 pKa = 10.64LSAFSWMYY15 pKa = 9.87EE16 pKa = 3.99QIFHH20 pKa = 7.21IISEE24 pKa = 4.46EE25 pKa = 4.14FPSKK29 pKa = 11.07YY30 pKa = 10.27YY31 pKa = 10.66DD32 pKa = 3.31NCCIIHH38 pKa = 6.9EE39 pKa = 4.57YY40 pKa = 10.42YY41 pKa = 10.12MSGTIIYY48 pKa = 9.55EE49 pKa = 4.05DD50 pKa = 4.06LFFDD54 pKa = 4.84GPHH57 pKa = 5.76KK58 pKa = 10.54RR59 pKa = 11.84IYY61 pKa = 10.85DD62 pKa = 3.66DD63 pKa = 4.17VINAGSAAYY72 pKa = 9.6YY73 pKa = 10.39RR74 pKa = 11.84KK75 pKa = 9.54FQALCFMVLGHH86 pKa = 6.9LEE88 pKa = 4.12PLFKK92 pKa = 10.81KK93 pKa = 10.14HH94 pKa = 6.31GDD96 pKa = 3.63KK97 pKa = 11.17LDD99 pKa = 3.66GLQIRR104 pKa = 11.84DD105 pKa = 3.83YY106 pKa = 11.19SSHH109 pKa = 7.24LDD111 pKa = 3.21QGLCYY116 pKa = 9.48IRR118 pKa = 11.84HH119 pKa = 6.19IFRR122 pKa = 11.84GYY124 pKa = 10.9LMEE127 pKa = 4.7RR128 pKa = 11.84MNFEE132 pKa = 4.01MANQYY137 pKa = 10.39ALKK140 pKa = 10.69SFDD143 pKa = 3.25SKK145 pKa = 10.61MVYY148 pKa = 8.25GTPSFEE154 pKa = 4.63FYY156 pKa = 10.58SKK158 pKa = 10.98SIMYY162 pKa = 9.43LQLKK166 pKa = 9.4HH167 pKa = 6.63ISTISDD173 pKa = 3.53SLKK176 pKa = 11.1DD177 pKa = 3.54GLKK180 pKa = 10.27SLKK183 pKa = 10.95SNDD186 pKa = 4.0LSDD189 pKa = 5.72DD190 pKa = 3.81DD191 pKa = 5.64DD192 pKa = 4.65EE193 pKa = 6.55SGEE196 pKa = 4.05ILKK199 pKa = 10.74KK200 pKa = 10.79LLLDD204 pKa = 3.92NTDD207 pKa = 3.25QTASTSCKK215 pKa = 9.81LKK217 pKa = 10.35RR218 pKa = 11.84QISIISIKK226 pKa = 10.67
Molecular weight: 26.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4B558|A0A0U4B558_9VIRU Uncharacterized protein OS=Actinidia chlorotic ringspot-associated virus OX=1776763 PE=4 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.16PMQGDD8 pKa = 3.79ASNSGSAVPSNVKK21 pKa = 9.68IDD23 pKa = 3.76PNTVKK28 pKa = 10.52FGKK31 pKa = 9.74KK32 pKa = 9.64NSVRR36 pKa = 11.84TITFTEE42 pKa = 4.45VVNNRR47 pKa = 11.84ATSSKK52 pKa = 10.37NSSNTAVSVFKK63 pKa = 11.14SPAFVGGFTSDD74 pKa = 3.71LDD76 pKa = 3.87FDD78 pKa = 3.45ITKK81 pKa = 9.57YY82 pKa = 10.77SRR84 pKa = 11.84VGSVEE89 pKa = 3.81TCASHH94 pKa = 7.32LSNSQEE100 pKa = 3.57IKK102 pKa = 10.41KK103 pKa = 10.46RR104 pKa = 11.84LVSDD108 pKa = 4.31GFLEE112 pKa = 4.79LPISNNYY119 pKa = 10.51SLFLCSEE126 pKa = 4.09IQATSIEE133 pKa = 4.26NAVSFNKK140 pKa = 10.1ACAIMAYY147 pKa = 9.9QILAHH152 pKa = 5.64TVMEE156 pKa = 4.61KK157 pKa = 10.87YY158 pKa = 10.3DD159 pKa = 3.54WEE161 pKa = 4.04QFKK164 pKa = 10.92AVEE167 pKa = 4.04VAIKK171 pKa = 10.21DD172 pKa = 3.63RR173 pKa = 11.84KK174 pKa = 9.74NPSISIVNRR183 pKa = 11.84LAGQMRR189 pKa = 11.84LPQSSLYY196 pKa = 10.04YY197 pKa = 8.86WMVVPGYY204 pKa = 10.09EE205 pKa = 3.77FLYY208 pKa = 10.67DD209 pKa = 3.42VFPIEE214 pKa = 4.45TIALTMVRR222 pKa = 11.84LEE224 pKa = 4.22CRR226 pKa = 11.84KK227 pKa = 10.04ALNIPEE233 pKa = 4.82TITNAEE239 pKa = 4.07IVNSLVSKK247 pKa = 9.54MNRR250 pKa = 11.84KK251 pKa = 9.02HH252 pKa = 6.93DD253 pKa = 3.78LTKK256 pKa = 10.82VSISEE261 pKa = 4.49IITKK265 pKa = 10.57LGVNNIKK272 pKa = 10.83DD273 pKa = 3.42MYY275 pKa = 11.09NEE277 pKa = 3.63FRR279 pKa = 11.84RR280 pKa = 11.84NVGTTGRR287 pKa = 11.84EE288 pKa = 3.6IRR290 pKa = 11.84NEE292 pKa = 3.88HH293 pKa = 5.89AVKK296 pKa = 10.51EE297 pKa = 4.01FLDD300 pKa = 5.32FIGKK304 pKa = 9.31YY305 pKa = 9.67CGSSTT310 pKa = 4.18
MM1 pKa = 7.84PKK3 pKa = 10.16PMQGDD8 pKa = 3.79ASNSGSAVPSNVKK21 pKa = 9.68IDD23 pKa = 3.76PNTVKK28 pKa = 10.52FGKK31 pKa = 9.74KK32 pKa = 9.64NSVRR36 pKa = 11.84TITFTEE42 pKa = 4.45VVNNRR47 pKa = 11.84ATSSKK52 pKa = 10.37NSSNTAVSVFKK63 pKa = 11.14SPAFVGGFTSDD74 pKa = 3.71LDD76 pKa = 3.87FDD78 pKa = 3.45ITKK81 pKa = 9.57YY82 pKa = 10.77SRR84 pKa = 11.84VGSVEE89 pKa = 3.81TCASHH94 pKa = 7.32LSNSQEE100 pKa = 3.57IKK102 pKa = 10.41KK103 pKa = 10.46RR104 pKa = 11.84LVSDD108 pKa = 4.31GFLEE112 pKa = 4.79LPISNNYY119 pKa = 10.51SLFLCSEE126 pKa = 4.09IQATSIEE133 pKa = 4.26NAVSFNKK140 pKa = 10.1ACAIMAYY147 pKa = 9.9QILAHH152 pKa = 5.64TVMEE156 pKa = 4.61KK157 pKa = 10.87YY158 pKa = 10.3DD159 pKa = 3.54WEE161 pKa = 4.04QFKK164 pKa = 10.92AVEE167 pKa = 4.04VAIKK171 pKa = 10.21DD172 pKa = 3.63RR173 pKa = 11.84KK174 pKa = 9.74NPSISIVNRR183 pKa = 11.84LAGQMRR189 pKa = 11.84LPQSSLYY196 pKa = 10.04YY197 pKa = 8.86WMVVPGYY204 pKa = 10.09EE205 pKa = 3.77FLYY208 pKa = 10.67DD209 pKa = 3.42VFPIEE214 pKa = 4.45TIALTMVRR222 pKa = 11.84LEE224 pKa = 4.22CRR226 pKa = 11.84KK227 pKa = 10.04ALNIPEE233 pKa = 4.82TITNAEE239 pKa = 4.07IVNSLVSKK247 pKa = 9.54MNRR250 pKa = 11.84KK251 pKa = 9.02HH252 pKa = 6.93DD253 pKa = 3.78LTKK256 pKa = 10.82VSISEE261 pKa = 4.49IITKK265 pKa = 10.57LGVNNIKK272 pKa = 10.83DD273 pKa = 3.42MYY275 pKa = 11.09NEE277 pKa = 3.63FRR279 pKa = 11.84RR280 pKa = 11.84NVGTTGRR287 pKa = 11.84EE288 pKa = 3.6IRR290 pKa = 11.84NEE292 pKa = 3.88HH293 pKa = 5.89AVKK296 pKa = 10.51EE297 pKa = 4.01FLDD300 pKa = 5.32FIGKK304 pKa = 9.31YY305 pKa = 9.67CGSSTT310 pKa = 4.18
Molecular weight: 34.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3871 |
226 |
2303 |
774.2 |
89.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.591 ± 0.571 | 1.937 ± 0.5 |
6.768 ± 0.444 | 5.399 ± 0.231 |
4.805 ± 0.375 | 3.565 ± 0.403 |
2.532 ± 0.416 | 8.835 ± 0.465 |
8.06 ± 0.258 | 9.429 ± 0.96 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.867 ± 0.361 | 6.071 ± 0.539 |
3.229 ± 0.194 | 2.532 ± 0.285 |
3.565 ± 0.23 | 9.326 ± 0.409 |
5.761 ± 0.308 | 5.296 ± 1.066 |
0.491 ± 0.103 | 5.942 ± 0.681 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
