
Blastochloris tepida
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Blastochloridaceae; Blastochloris
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
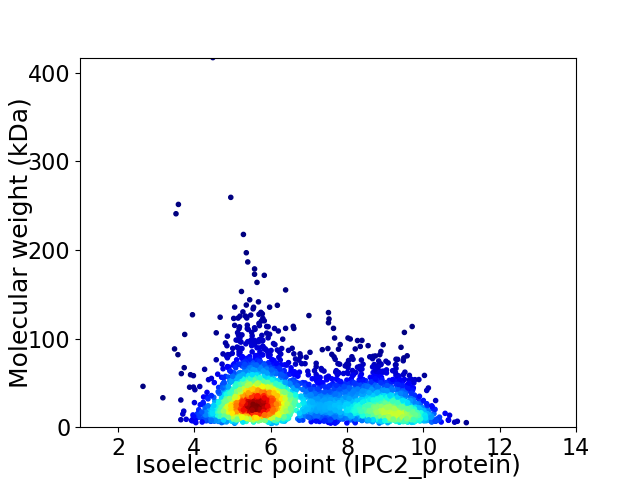
Virtual 2D-PAGE plot for 3554 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A348FZ47|A0A348FZ47_9RHIZ Uncharacterized protein OS=Blastochloris tepida OX=2233851 GN=BLTE_12650 PE=4 SV=1
MM1 pKa = 7.61NIHH4 pKa = 7.28DD5 pKa = 3.98VTEE8 pKa = 5.01LIWHH12 pKa = 6.29FVGQLRR18 pKa = 11.84LDD20 pKa = 3.84EE21 pKa = 4.86DD22 pKa = 4.25LSRR25 pKa = 11.84SRR27 pKa = 11.84IQYY30 pKa = 9.73EE31 pKa = 3.76DD32 pKa = 3.8AEE34 pKa = 4.44AGRR37 pKa = 11.84VRR39 pKa = 11.84EE40 pKa = 4.15DD41 pKa = 2.99VFGRR45 pKa = 11.84VKK47 pKa = 10.51DD48 pKa = 3.59APVRR52 pKa = 11.84VEE54 pKa = 5.08DD55 pKa = 4.63PGEE58 pKa = 4.15SAPTPVSYY66 pKa = 9.13ATFAAAPALTPFVPHH81 pKa = 6.61FMQPGLASVPVEE93 pKa = 4.16PLDD96 pKa = 5.35DD97 pKa = 3.26IHH99 pKa = 7.01FKK101 pKa = 10.81VPPRR105 pKa = 11.84VLAAPAPGSLAGWYY119 pKa = 6.78PQSIGPEE126 pKa = 3.77YY127 pKa = 10.57HH128 pKa = 5.16QTLFEE133 pKa = 4.3ARR135 pKa = 11.84QINQLNDD142 pKa = 2.99SDD144 pKa = 4.61IFSDD148 pKa = 3.82DD149 pKa = 3.99PLWAVALPVAKK160 pKa = 7.74TTWSVDD166 pKa = 3.15EE167 pKa = 5.31LYY169 pKa = 9.91ATAQDD174 pKa = 5.07AIPAHH179 pKa = 6.78LEE181 pKa = 4.02TVAGGGSQEE190 pKa = 4.44AIAYY194 pKa = 6.66ATSLGSGAYY203 pKa = 9.41AFPSSQVEE211 pKa = 3.85PGSYY215 pKa = 10.52HH216 pKa = 5.4NGEE219 pKa = 4.6RR220 pKa = 11.84LAEE223 pKa = 4.39DD224 pKa = 3.08QTLPTDD230 pKa = 3.48IGATVPEE237 pKa = 4.77MPQPTNGPTGDD248 pKa = 4.55LGITTQAAEE257 pKa = 4.21LGANGAYY264 pKa = 10.19NSALIADD271 pKa = 4.27ANGACGTLVVLGDD284 pKa = 4.05KK285 pKa = 10.76FSLNAIIQINAYY297 pKa = 10.01SDD299 pKa = 3.5SDD301 pKa = 4.09VINAYY306 pKa = 10.06GGAGYY311 pKa = 8.08GTAGTIAGGGNQADD325 pKa = 4.0NLAQVGFTEE334 pKa = 4.77LEE336 pKa = 4.03AMPSAKK342 pKa = 10.2SGTPYY347 pKa = 10.55RR348 pKa = 11.84IDD350 pKa = 3.41VVEE353 pKa = 4.78GDD355 pKa = 3.75FFDD358 pKa = 5.22IRR360 pKa = 11.84TLYY363 pKa = 10.32QSNTLLDD370 pKa = 3.58SDD372 pKa = 4.43GASLTTEE379 pKa = 4.01GHH381 pKa = 6.06FSLVRR386 pKa = 11.84TGQNEE391 pKa = 4.41LVNLADD397 pKa = 5.64LSDD400 pKa = 4.44FDD402 pKa = 4.55LFSYY406 pKa = 10.95YY407 pKa = 10.75DD408 pKa = 3.51VIVIGGQYY416 pKa = 10.58YY417 pKa = 10.3DD418 pKa = 3.57INAIIQLNLLNDD430 pKa = 3.65NDD432 pKa = 3.97AVSAGGCGAGSGAGAGISTGGNSLLNDD459 pKa = 3.54ARR461 pKa = 11.84IEE463 pKa = 4.25NYY465 pKa = 8.03GTTNVQPLDD474 pKa = 3.57DD475 pKa = 5.26DD476 pKa = 3.81MAKK479 pKa = 10.61FSASLADD486 pKa = 4.18GDD488 pKa = 4.67APDD491 pKa = 5.01GASSWGFSDD500 pKa = 5.43GGDD503 pKa = 3.54GYY505 pKa = 10.75IDD507 pKa = 3.47VLYY510 pKa = 9.86VTGNFFDD517 pKa = 5.97INLLWQINTVTDD529 pKa = 3.0VDD531 pKa = 3.94AAALQAGTDD540 pKa = 3.78GSATQVVSTGANEE553 pKa = 4.28LQNQAIIVDD562 pKa = 3.95VDD564 pKa = 3.56ALTSEE569 pKa = 4.69YY570 pKa = 10.33IGGDD574 pKa = 3.43VYY576 pKa = 11.08EE577 pKa = 4.56GSILIQANLVEE588 pKa = 4.49SDD590 pKa = 3.39EE591 pKa = 4.98DD592 pKa = 4.17SVVHH596 pKa = 6.94HH597 pKa = 6.73DD598 pKa = 3.85TEE600 pKa = 4.57TLVPEE605 pKa = 4.8VIAFTGGSDD614 pKa = 4.05DD615 pKa = 5.32CGHH618 pKa = 7.11TDD620 pKa = 3.33PTYY623 pKa = 10.41VGPPVTMTDD632 pKa = 3.83HH633 pKa = 7.23ILGTVMTT640 pKa = 5.17
MM1 pKa = 7.61NIHH4 pKa = 7.28DD5 pKa = 3.98VTEE8 pKa = 5.01LIWHH12 pKa = 6.29FVGQLRR18 pKa = 11.84LDD20 pKa = 3.84EE21 pKa = 4.86DD22 pKa = 4.25LSRR25 pKa = 11.84SRR27 pKa = 11.84IQYY30 pKa = 9.73EE31 pKa = 3.76DD32 pKa = 3.8AEE34 pKa = 4.44AGRR37 pKa = 11.84VRR39 pKa = 11.84EE40 pKa = 4.15DD41 pKa = 2.99VFGRR45 pKa = 11.84VKK47 pKa = 10.51DD48 pKa = 3.59APVRR52 pKa = 11.84VEE54 pKa = 5.08DD55 pKa = 4.63PGEE58 pKa = 4.15SAPTPVSYY66 pKa = 9.13ATFAAAPALTPFVPHH81 pKa = 6.61FMQPGLASVPVEE93 pKa = 4.16PLDD96 pKa = 5.35DD97 pKa = 3.26IHH99 pKa = 7.01FKK101 pKa = 10.81VPPRR105 pKa = 11.84VLAAPAPGSLAGWYY119 pKa = 6.78PQSIGPEE126 pKa = 3.77YY127 pKa = 10.57HH128 pKa = 5.16QTLFEE133 pKa = 4.3ARR135 pKa = 11.84QINQLNDD142 pKa = 2.99SDD144 pKa = 4.61IFSDD148 pKa = 3.82DD149 pKa = 3.99PLWAVALPVAKK160 pKa = 7.74TTWSVDD166 pKa = 3.15EE167 pKa = 5.31LYY169 pKa = 9.91ATAQDD174 pKa = 5.07AIPAHH179 pKa = 6.78LEE181 pKa = 4.02TVAGGGSQEE190 pKa = 4.44AIAYY194 pKa = 6.66ATSLGSGAYY203 pKa = 9.41AFPSSQVEE211 pKa = 3.85PGSYY215 pKa = 10.52HH216 pKa = 5.4NGEE219 pKa = 4.6RR220 pKa = 11.84LAEE223 pKa = 4.39DD224 pKa = 3.08QTLPTDD230 pKa = 3.48IGATVPEE237 pKa = 4.77MPQPTNGPTGDD248 pKa = 4.55LGITTQAAEE257 pKa = 4.21LGANGAYY264 pKa = 10.19NSALIADD271 pKa = 4.27ANGACGTLVVLGDD284 pKa = 4.05KK285 pKa = 10.76FSLNAIIQINAYY297 pKa = 10.01SDD299 pKa = 3.5SDD301 pKa = 4.09VINAYY306 pKa = 10.06GGAGYY311 pKa = 8.08GTAGTIAGGGNQADD325 pKa = 4.0NLAQVGFTEE334 pKa = 4.77LEE336 pKa = 4.03AMPSAKK342 pKa = 10.2SGTPYY347 pKa = 10.55RR348 pKa = 11.84IDD350 pKa = 3.41VVEE353 pKa = 4.78GDD355 pKa = 3.75FFDD358 pKa = 5.22IRR360 pKa = 11.84TLYY363 pKa = 10.32QSNTLLDD370 pKa = 3.58SDD372 pKa = 4.43GASLTTEE379 pKa = 4.01GHH381 pKa = 6.06FSLVRR386 pKa = 11.84TGQNEE391 pKa = 4.41LVNLADD397 pKa = 5.64LSDD400 pKa = 4.44FDD402 pKa = 4.55LFSYY406 pKa = 10.95YY407 pKa = 10.75DD408 pKa = 3.51VIVIGGQYY416 pKa = 10.58YY417 pKa = 10.3DD418 pKa = 3.57INAIIQLNLLNDD430 pKa = 3.65NDD432 pKa = 3.97AVSAGGCGAGSGAGAGISTGGNSLLNDD459 pKa = 3.54ARR461 pKa = 11.84IEE463 pKa = 4.25NYY465 pKa = 8.03GTTNVQPLDD474 pKa = 3.57DD475 pKa = 5.26DD476 pKa = 3.81MAKK479 pKa = 10.61FSASLADD486 pKa = 4.18GDD488 pKa = 4.67APDD491 pKa = 5.01GASSWGFSDD500 pKa = 5.43GGDD503 pKa = 3.54GYY505 pKa = 10.75IDD507 pKa = 3.47VLYY510 pKa = 9.86VTGNFFDD517 pKa = 5.97INLLWQINTVTDD529 pKa = 3.0VDD531 pKa = 3.94AAALQAGTDD540 pKa = 3.78GSATQVVSTGANEE553 pKa = 4.28LQNQAIIVDD562 pKa = 3.95VDD564 pKa = 3.56ALTSEE569 pKa = 4.69YY570 pKa = 10.33IGGDD574 pKa = 3.43VYY576 pKa = 11.08EE577 pKa = 4.56GSILIQANLVEE588 pKa = 4.49SDD590 pKa = 3.39EE591 pKa = 4.98DD592 pKa = 4.17SVVHH596 pKa = 6.94HH597 pKa = 6.73DD598 pKa = 3.85TEE600 pKa = 4.57TLVPEE605 pKa = 4.8VIAFTGGSDD614 pKa = 4.05DD615 pKa = 5.32CGHH618 pKa = 7.11TDD620 pKa = 3.33PTYY623 pKa = 10.41VGPPVTMTDD632 pKa = 3.83HH633 pKa = 7.23ILGTVMTT640 pKa = 5.17
Molecular weight: 67.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A348G409|A0A348G409_9RHIZ TIGR02300 family protein OS=Blastochloris tepida OX=2233851 GN=BLTE_29770 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84KK29 pKa = 9.21VIAARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.21RR41 pKa = 11.84LSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84KK29 pKa = 9.21VIAARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.21RR41 pKa = 11.84LSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1106585 |
41 |
4128 |
311.4 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.196 ± 0.069 | 0.827 ± 0.014 |
5.678 ± 0.032 | 5.7 ± 0.046 |
3.494 ± 0.026 | 8.592 ± 0.046 |
1.921 ± 0.022 | 4.692 ± 0.032 |
2.946 ± 0.04 | 10.11 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.178 ± 0.021 | 2.16 ± 0.025 |
5.712 ± 0.048 | 2.773 ± 0.022 |
7.921 ± 0.059 | 4.783 ± 0.035 |
5.287 ± 0.039 | 7.831 ± 0.033 |
1.264 ± 0.017 | 1.934 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
